Categories of Database Technical Design Data source Data
Categories of Database -Technical Design -Data source -Data access -Any other parameter
Technical Design • • Flat File XML Relational model ASN. 1
Flat File • The simplest database model • Information is stored in text file, one record per line format • Each record represents one entry having field of a fixed width or fields separated by delimiters e. g. white space, tabs, commas or other character • Supports simple text files that can be accessed easily by simple programs to handle a query
• Exists in a single file consisting of data and delimiters in the form of rows and columns • Analogy is a list of contacts comprising names, addresses and phone nos • Are indexed for easier search • 2 common file formats supported the flat files are FASTA format (free text) and EMBL format (tag, value pairs)
FASTA Format • In the process of writing a similarity searching program (in 1985), William Pearson designed a simple text format for DNA and protein sequences • The FASTA format is now universal for all databases and software that handles DNA and protein sequences
![• One header line, starts with > with a [return] at end • • One header line, starts with > with a [return] at end •](http://slidetodoc.com/presentation_image_h2/6d2b96b059c468acf73e964b30966c3d/image-6.jpg)
• One header line, starts with > with a [return] at end • All other characters are part of sequence. Most software ignores spaces, carriage returns. Some ignores numbers >URO 1 uro 1. seq N Check: 3854 Length: 2018. . November 9, 2000 11: 50 Type: CGCAGAAAGAGGAGGCGCTTGCCTTCAGCTTGTGGGAAATCCCGAAGATGGCC AAAGACAACTGTTCGTTGCTTCCAGGGCCTGCTGATTTTTGGAAATGT GATTATTGGTTGTTGCGGCATTGCCCTGACTGCGGAGTGCATCTTCTTTGTAT CTGACCAACACAGCCTCTACC CACTGCTTGAAGCCACCGACAACGATGACATCTATGGGGCTGCCTGGATCGGC ATATTTGTGGGCATCTGCCTCTTCTGCCTGTTCTAGGCATTGTAGGCAT CATGAAGTCCAGCAGGAAAATTCTTCTGGCGTATTTCATTCTGATGTTTATAG TATATGCCTTTGAAGTGGCATCTTGTATCACAGCAGCAACAAGACTTT TTCACACCCAACCTCTTCCTGAAGCAGATGCTAGAGAGGTACCAAAACAACAG CCCTCCAAACAATGATGACCAGTGGAAAAACAATG

Ctd’. . other example… • >gi 1040960 gb U 35641. 1 MMU 35641 Mus musculus Brcal 1 m. RNA, complete cds • CGTAGGGCGCGGATAGCGATCG GATTTCCGAGGAGATTATAAAGATT AGAGGGACTTTTAAGTAGGAG CCCCCTCTCTCGACACCAAACGG

Multi-Sequence FASTA file >FBpp 0074027 type=protein; loc=X: complement(16159413. . 16159860, 16160061. . 16160497); ID=FBpp 0074027; name=CG 12507 -PA; parent=FBgn 0030729, FBtr 0074248; dbxref=Fly. Base: FBpp 0074027, Fly. Base_Annotation_IDs: CG 12507 PA, GB_protein: AAF 48569. 1, GB_protein: AAF 48569; MD 5=123 b 97 d 79 d 04 a 06 c 66 e 12 fa 665 e 6 d 801; release=r 5. 1; species=Dmel; length=294; MRCLMPLLLANCIAANPSFEDPDRSLDMEAKDSSVVDTMGMGMGVLDPTQ PKQMNYQKPPLGYKDYDYYLGSRRMADPYGADNDLSASSAIKIHGEGNLA SLNRPVSGVAHKPLPWYGDYSGKLLASAPPMYPSRSYDPYIRRYDRYDEQ YHRNYPQYFEDMYMHRQRFDPYDSYSPRIPQYPEPYVMYPDRYPDAPPLR DYPKLRRGYIGEPMAPIDSYSSSKYVSSKQSDLSFPVRNERIVYYAHLPE IVRTPYDSGSPEDRNSAPYKLNKKKIKNIQRPLANNSTTYKMTL >FBpp 0082232 type=protein; loc=3 R: complement(9207109. . 9207225, 9207285. . 9207431); ID=FBpp 0082232; name=m. Rp. S 21 -PA; parent=FBgn 0044511, FBtr 0082764; dbxref=Fly. Base: FBpp 0082232, Fly. Base_Annotation_IDs: CG 32854 PA, GB_protein: AAN 13563. 1, GB_protein: AAN 13563; MD 5=dcf 91821 f 75 ffab 320491 d 124 a 0 d 816 c; release=r 5. 1; species=Dmel; length=87; MRHVQFLARTVLVQNNNVEEACRLLNRVLGKEELLDQFRRTRFYEKPYQV RRRINFEKCKAIYNEDMNRKIQFVLRKNRAEPFPGCS >FBpp 0091159 type=protein; loc=2 R: complement(2511337. . 2511531, 2511594. . 2511767, 2511824. . 2511979, 2512032. . 2512082); ID=FBpp 0091159; name=CG 33919 -PA; parent=FBgn 0053919, FBtr 0091923; dbxref=Fly. Base: FBpp 0091159, Fly. Base_Annotation_IDs: CG 33919 -PA, GB_protein: AAZ 52801. 1, GB_protein: AAZ 52801; MD 5=c 91 d 880 b 654 cd 612 d 7292676 f 95038 c 5; release=r 5. 1; species=Dmel; length=191; MKLVLVVLLGCCFIGQLTNTQLVYKLKKIECLVNRTRVSNVSCHVKAINWNLAVVNMDCFMIVPLHNPIIRMQVFTKDYSNQYKPFLVDVKIRICEVIE RRNFIPYGVIMWKLFKRYTNVNHSCPFSGHLIARDGFLDTSLLPPFPQGFYQVSLVVTDTNSTSTDYVGTMKFFLQAMEHIKSKKTHNLVHN >FBpp 0070770 type=protein; loc=X: join(5584802. . 5585021, 5585925. . 5586137, 5586198. . 5586342, 5586410. . 5586605); ID=FBpp 0070770; name=cv-PA; parent=FBgn 0000394, FBtr 0070804; dbxref=Fly. Base: FBpp 0070770, Fly. Base_Annotation_IDs: CG 12410 PA, GB_protein: AAF 46063. 1, GB_protein: AAF 46063; MD 5=0626 ee 34 a 518 f 248 bbdda 11 a 211 f 9 b 14; release=r 5. 1; species=Dmel; length=257; MEIWRSLTVGTIVLLAIVCFYGTVESCNEVVCASIVSKCMLTQSCKCELKNCSCCKECLKCLGKNYEECCSCVELCPKPNDTRNSLSKKSHVEDFDGVPE LFNAVATPDEGDSFGYNWNVFTFQVDFDKYLKGPKLEKDGHYFLRTNDKNLDEAIQERDNIVTVNCTVIYLDQCVSWNKCRTSCQTTGASSTRWFH DGCCECVGSTCINYGVNESRCRKCPESKGELGDELDDPMEEEMQDFGESMGPFD GPVNNNY …
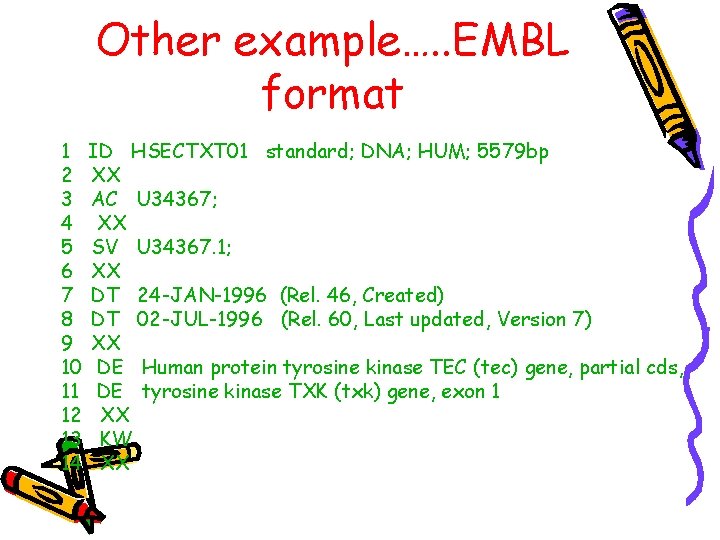
Other example…. . EMBL format 1 ID HSECTXT 01 standard; DNA; HUM; 5579 bp 2 XX 3 AC U 34367; 4 XX 5 SV U 34367. 1; 6 XX 7 DT 24 -JAN-1996 (Rel. 46, Created) 8 DT 02 -JUL-1996 (Rel. 60, Last updated, Version 7) 9 XX 10 DE Human protein tyrosine kinase TEC (tec) gene, partial cds, 11 DE tyrosine kinase TXK (txk) gene, exon 1 12 XX 13 KW 14 XX
XML File • Type hierarchical and semi-structured model • has text-based in the form of e. Xtensible Markup Language • Support XQuery as the query language • Nested data structures that has implicit, flexible schema

• • • Example: XML file format <xml version=“ 1. 0> <!DOCTYPE GBSeq PUBLIC”-//NCBI GBSeq/EN” http: //www. ncbi. nlm. nih. gov/dtd/NCBI_GBSeq. dtd> <GBSeq> <GBSeq_locus>MMU 35641</GBSeq_locus> <GBSeq_Length>5538<GBSeq_Length> <GBSeq_strandedness values=“notset>0</GBSeq_strandedness> <GBSeq_moltype value=“mrna”>5</GBSeq_moltype> <GBSeq_topology value= “linear” >1</GBSeq_topology> <GBSeq_division>ROD</GBSeq_division <GBSeq_update-date>18 -OCT_1996</GBSeq_update-date>

Relational File • A relational database stores information in tables. • Each inforamtion theme is stored in its own table • Each table is named and contain rows called tuples or record and column called attributes or fields • A combination of attributes is called a key that identify each record • In essence, a relational database will break-up a list into several parts. – One part for each theme in the list – For example, this might be divided with the author, the jurnal title and some definations about the gene or protein
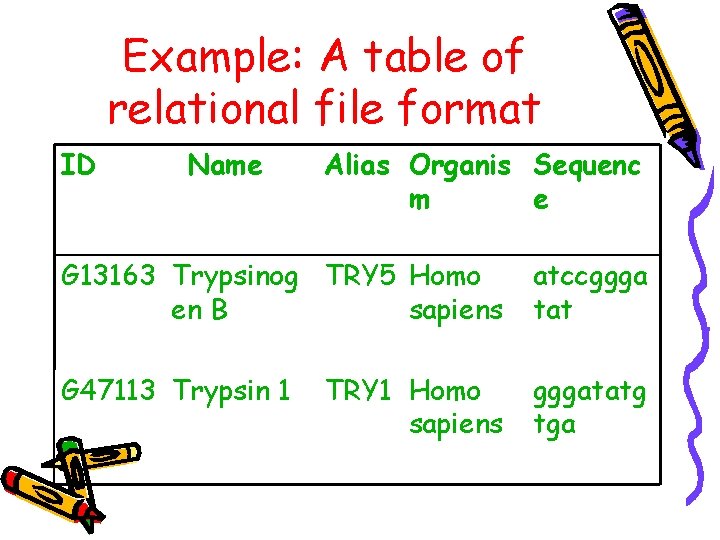
Example: A table of relational file format ID Name Alias Organis Sequenc m e G 13163 Trypsinog TRY 5 Homo en B sapiens atccggga tat G 47113 Trypsin 1 gggatatg tga TRY 1 Homo sapiens

• A relational database is more complicated than a list • However, a relational database minimises data redundancy, preserves complex realtionships among topics and allows for partial data • Futhermore, a realtional database provides a solid for creating user interface forms and reports

ASN. 1 File • Abstract Syntax Notation one • It is a notation used for representing data to communicate information across various platforms • Some understand it as a language that is used to define information so that it can be ported over the internet • An application protocol to represent data format

• It has 2 basic kinds of syntax: i). the abstract syntax- to define information ii) the transfer syntax – to communicate information • It has structured flat-file format • Supports multiple data types
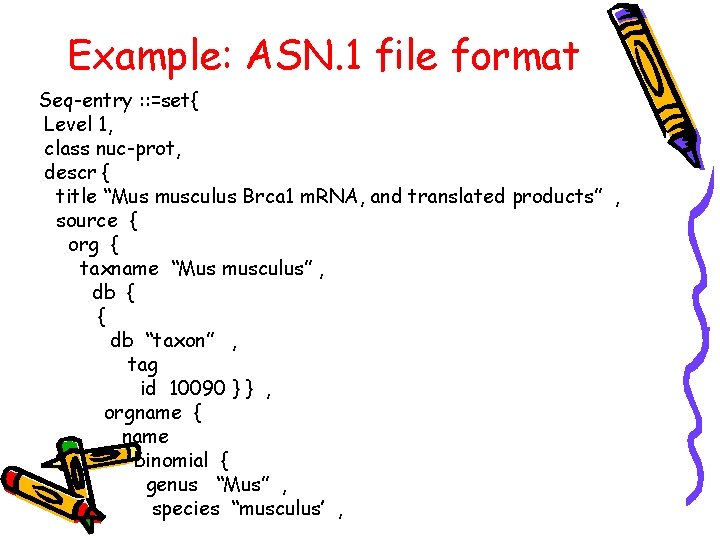
Example: ASN. 1 file format Seq-entry : : =set{ Level 1, class nuc-prot, descr { title “Mus musculus Brca 1 m. RNA, and translated products” , source { org { taxname “Mus musculus” , db { { db “taxon” , tag id 10090 } } , orgname { name binomial { genus “Mus” , species “musculus’ ,
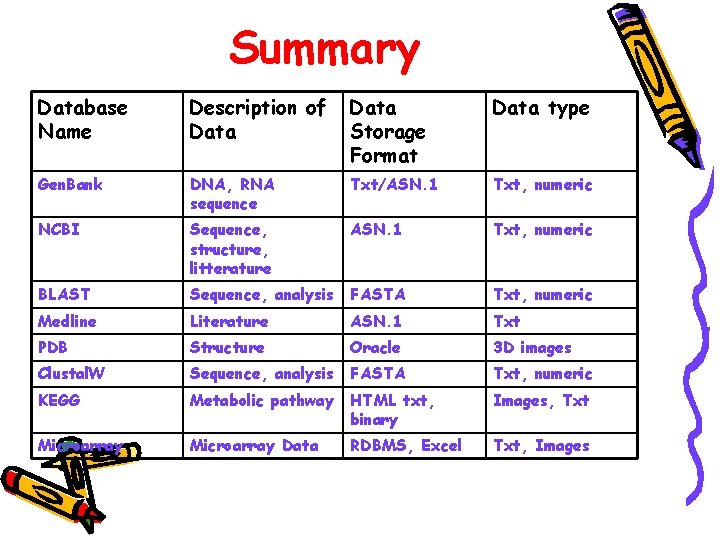
Summary Database Name Description of Data Storage Format Data type Gen. Bank DNA, RNA sequence Txt/ASN. 1 Txt, numeric NCBI Sequence, structure, litterature ASN. 1 Txt, numeric BLAST Sequence, analysis FASTA Txt, numeric Medline Literature ASN. 1 Txt PDB Structure Oracle 3 D images Clustal. W Sequence, analysis FASTA Txt, numeric KEGG Metabolic pathway HTML txt, binary Images, Txt Microarray Data RDBMS, Excel Txt, Images
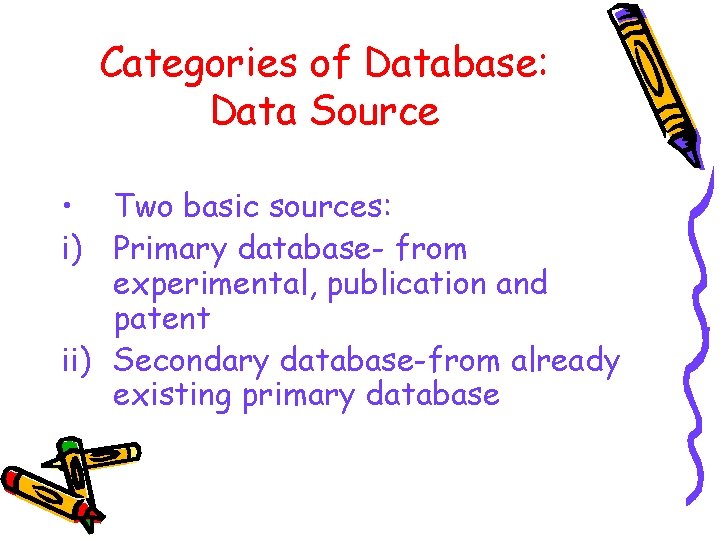
Categories of Database: Data Source • Two basic sources: i) Primary database- from experimental, publication and patent ii) Secondary database-from already existing primary database

Categories of Database: Data Access • Publicly available with no restrictions, such as databases of NCBI, EMBL and SWISS-PROT • Available with copyright • Browsing only, accessible but not downloadable • Academic, but not freely available • Proprietary, commercial with access on payment, e. g. PUBMED and MEDLINE • Ad-Hoc query facility, e. g. Boolean queries • Unrestricted SQL queries against underlying DBMS
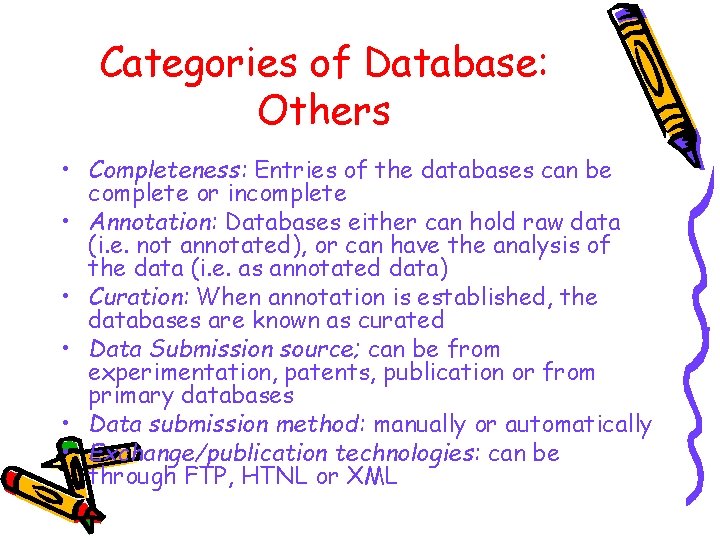
Categories of Database: Others • Completeness: Entries of the databases can be complete or incomplete • Annotation: Databases either can hold raw data (i. e. not annotated), or can have the analysis of the data (i. e. as annotated data) • Curation: When annotation is established, the databases are known as curated • Data Submission source; can be from experimentation, patents, publication or from primary databases • Data submission method: manually or automatically • Exchange/publication technologies: can be through FTP, HTNL or XML
Structured Query Language (SQL) • Is an international standard languange for creating, processing and querying databases and their table • The vast majority of data-driven applications and websites use SQL to retrieve, format, report insert, delete and / or modify data for users.
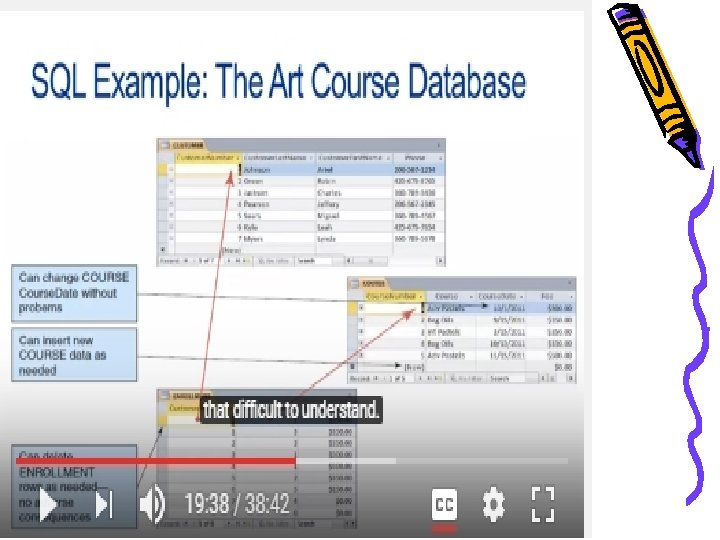
SQL example • We can use SQL to combine the data in the three tables to create the original list structure of the data • We do this by using a SQL SELECT statement
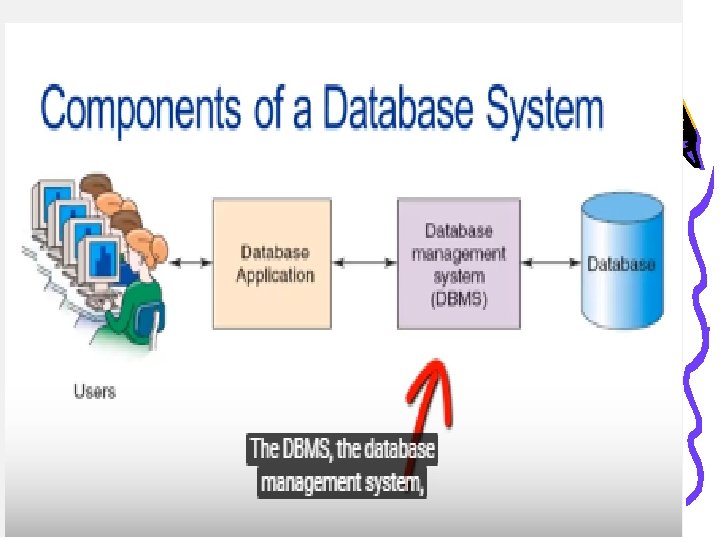
Database System • The four components of a database system are: – – Users Database Application (s) Database Management System (DBMS) Database
Users • A user of a database system will: – Use a database application to keep track of information – Use different user interface forms to enter, read, delete and query data – Produce reports

The Database • Is a self-describing collection related records – Self-describing: • The database itself contains the defination of its structure • Metadata are data describe the strudture of the data in Tables within a relational database are related to each other in some way
Database Management System (DBMS) • Serves as an intermediary between database applications and the database • DBMS manages and controls database activities • The DBMS creates, processes and administers the databases it controls
• • • Functions of DBMS Creates databases Create tables Create supporting structures Read database data Modify database data (insert, update, delete) • Enforce rules • Provide security
Database applications • Is a set of one or more computer programs or websites that serve as an intermediary between the user and the DBMS
- Slides: 31