Capillary Electrophoresis CE Dr Muhammad Shafique Forensic DNA











- Slides: 29

Capillary Electrophoresis (CE) Dr. Muhammad Shafique Forensic DNA Scientist CEMB, Lahore 1

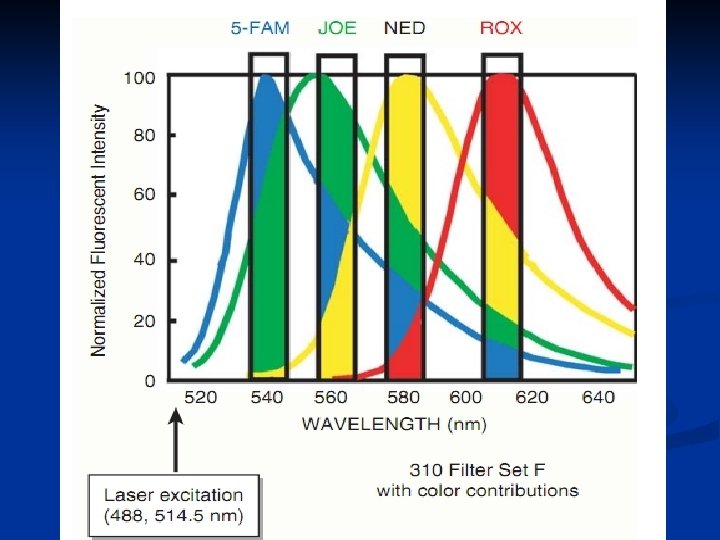
Since the late 1980 s, methods such as silver staining and fluorescence techniques become popular for detecting short tandem repeat (STR) alleles due to their low cost in the case of silver staining and their capability of automating the detection in the case of fluorescence. The first CE separations of DNA were performed in the late 1980 s. Since the introduction of new CE instrumentation in the mid 1990 s, the technique has gained rapidly in popularity. Fluorescence Detection Fluorescence measurements involve exciting a dye molecule and then detecting the light that is emitted from the excited dye. A molecule that is capable of fluorescence is called a fluorophore. Fluorophores come in a variety of shapes, sizes, and abilities. The ones that are primarily used in DNA labeling are dyes that fluoresce in the visible region of the spectrum, which consists of light emitted in the range of approximately 400– 600 nm. December 15, 2021 CEMB, Lahore 2

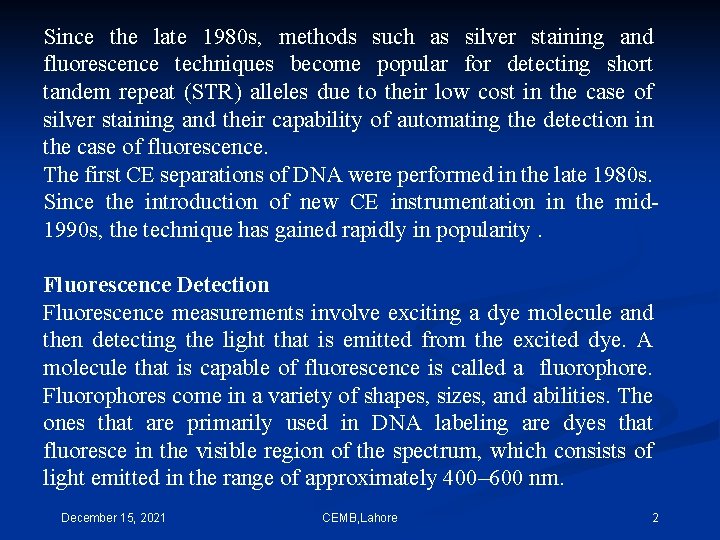
In the first step, a photon (hνex) from a laser source excites a fluorophore electron from its ground energy state (S 0) to an excited transition state (S′ 1). This electron then undergoes conformational changes and interacts with its environment resulting in the relaxed singlet excitation state (S 1). During the final step of the process, a photon (hνem) is emitted at a lower energy when the excited electron falls back to its ground state. Because energy and wavelength are inversely related to one another, the emission photon has a higher wavelength than the excitation photon. The difference between the apex of the absorption and emission spectra is called the Stokes shift. This shift permits the use of optical filters to separate excitation light from emission light. Fluorophores have characteristic light absorption and emission patterns that are based upon their chemical structure and the environmental conditions. With careful selection and optical filters, fluorophores may be chosen with emission spectra that are resolvable from one another. December 15, 2021 CEMB, Lahore 3

December 15, 2021 CEMB, Lahore 4

Factors that affect how well a fluorophore will emit light, or fluoresce. • Molar extinction coefficient: the ability of a dye to absorb light; • Quantum yield: the efficiency with which the excited fluorophore converts absorbed light to emitted light. • Photo stability: the ability of a dye to undergo repeated cycles of excitation and emission without being destroyed in the excited state, or experiencing ‘photobleaching’ • Dye environment: factors that affect fluorescent yield include p. H, temperature, solvent and the presence of quenchers, such as hemoglobin. December 15, 2021 CEMB, Lahore 5

The intensity of the light emitted by a fluorophore is directly dependent on the amount of light that the dye has absorbed. Lasers are an effective excitation source because the light they emit is very intense and at primarily one wavelength. Argon ion gas laser (Ar+): produces light at 488 nm and 514. 5 nm and most popular for applications involving fluorescent DNA labeling because a number of dyes are available that closely match its excitation capabilities. Solid-state Nd: YAG laser : produces a beam of light at 532 nm December 15, 2021 CEMB, Lahore 6

A fluorescence detector is a photosensitive device that measures the light intensity emitted from a fluorophore. Detection of low-intensity light may be accomplished with a photomultiplier tube (PMT) or a charge-coupled device (CCD). In both cases, the action of a photon striking the detector is converted to an electric signal. The strength of the resultant current is proportional to the intensity of the incident light. This light intensity is typically reported in arbitrary units, such as relative fluorescence units (RFUs). December 15, 2021 CEMB, Lahore 7

December 15, 2021 CEMB, Lahore 8

December 15, 2021 CEMB, Lahore 9

December 15, 2021 CEMB, Lahore 10

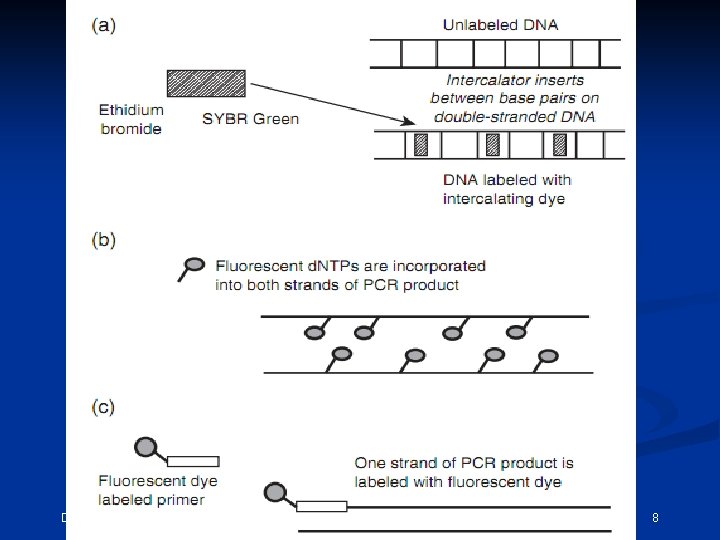
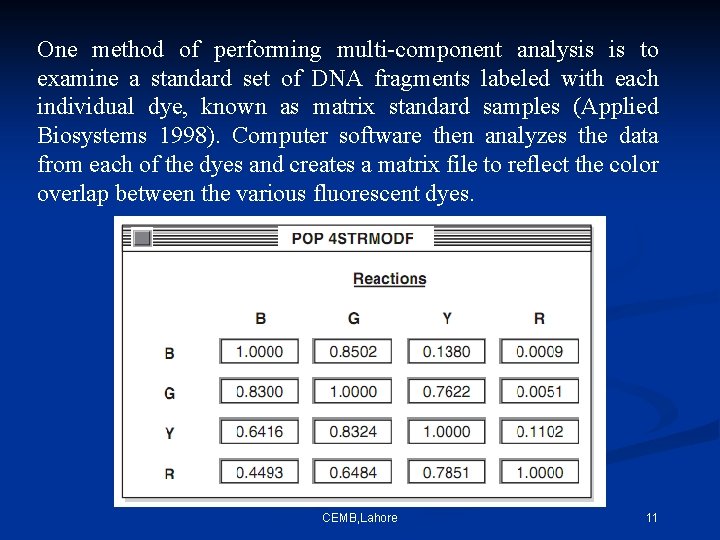
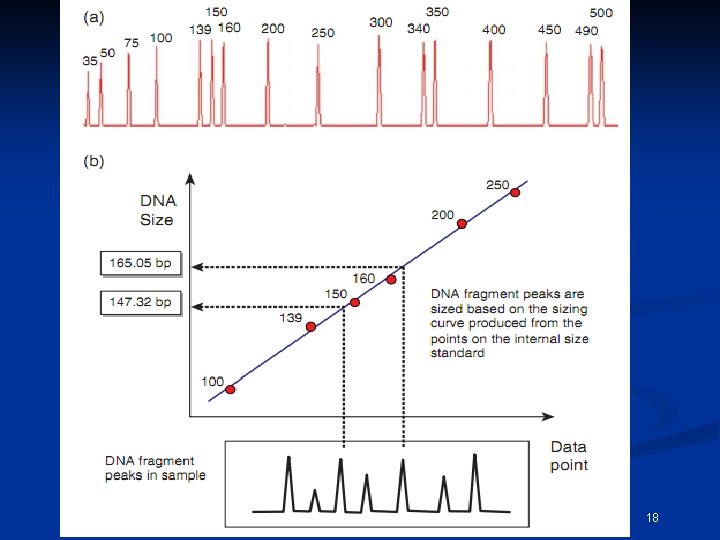
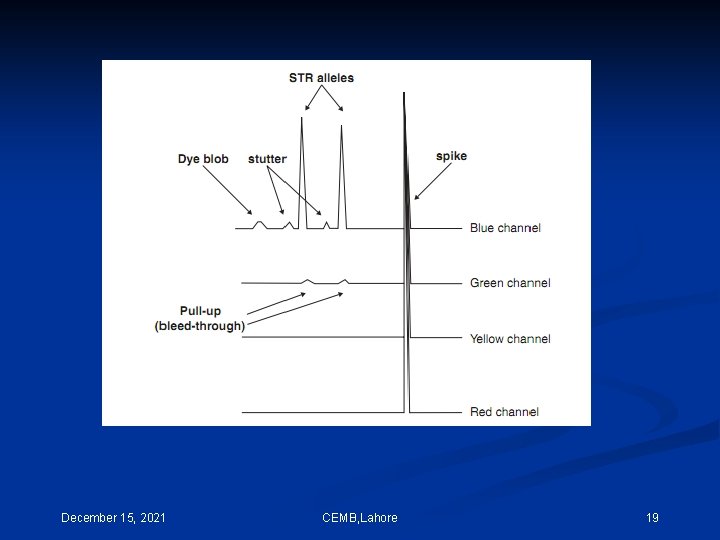
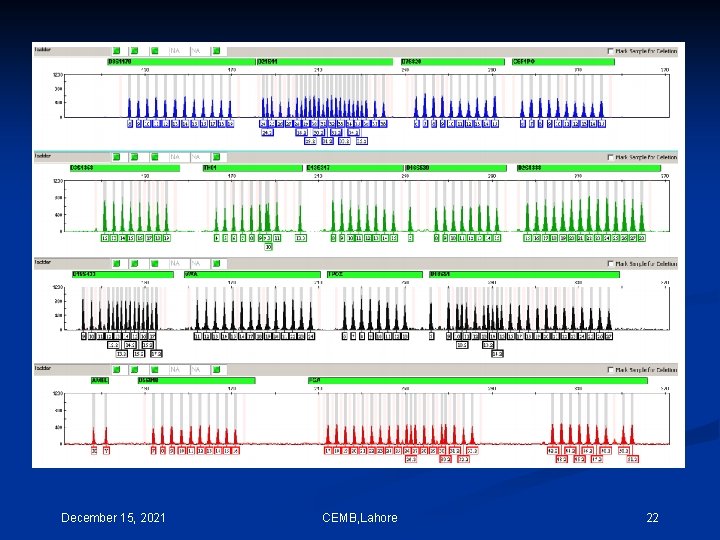
One method of performing multi-component analysis is to examine a standard set of DNA fragments labeled with each individual dye, known as matrix standard samples (Applied Biosystems 1998). Computer software then analyzes the data from each of the dyes and creates a matrix file to reflect the color overlap between the various fluorescent dyes. CEMB, Lahore 11

If the matrix color deconvolution (algorithm-based signal process ) does not work properly than the baseline can be uneven or a phenomenon known as ‘pull-up’ can occur. Pull-up is the result of a color bleeding from one spectral channel into another, usually because of off-scale peaks. The most common occurrence of pullup involves small green peaks showing up under blue peaks that are off-scale. This occurs because of the significant overlap of the blue and green dyes. December 15, 2021 CEMB, Lahore 12

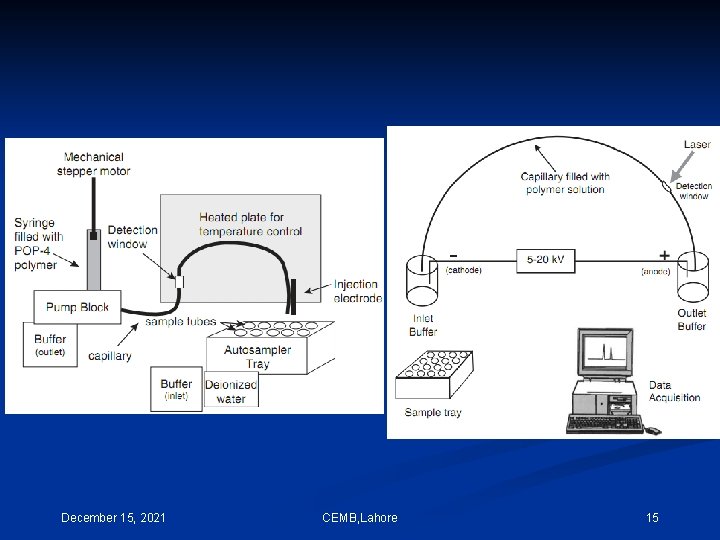
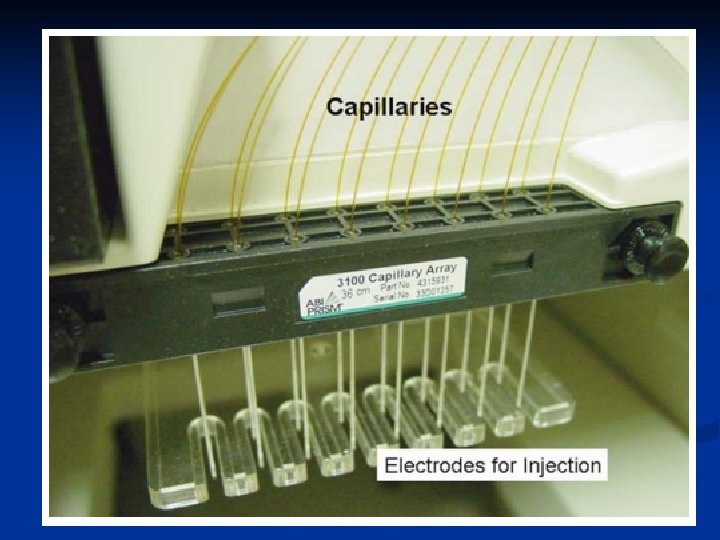
Typical DNA separation times using CE are in the range of 5– 30 minutes. Electrokinetic injection capillary length (ABI 310) = 47 cm Separation voltage = 15 KV or 320 V/cm Capillary have an inner diameter = 50, 75, or 100 µm for STR separations Capillary diameter = 50 µm Polymer Solution: POP-4™ and POP-6™ (Performance Optimized Polymer) 4% and 6% concentrations of linear, uncross-linked dimethyl polyacrylamide, respectively. A high concentration of urea is also present in the polymer solution to create an environment in the capillary that will keep the DNA molecules denatured. POP-4 is a commercially available preparation of a flowable polymer [poly (N, N-dimethylacrylamide); DMA]. POP-4 contains 4% DMA homo-polymer, 8 M urea, 5% 2 -pyrrolidinone, and 100 m. M N-Tris-(hydroxymethyl) methyl-3 -aminopropane-sulfonic acid (TAPS) adjusted to p. H 8. 0 with Na. OH December 15, 2021 CEMB, Lahore 13

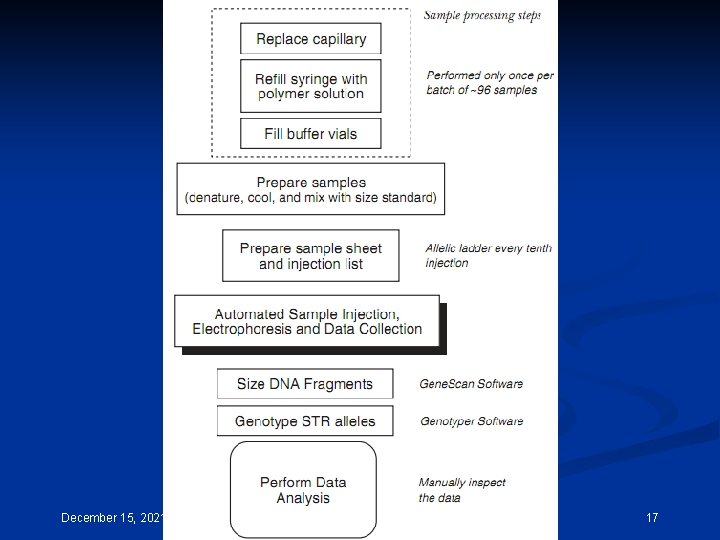
POP-4 is most commonly used for STR typing while POP-6, which is more viscous and yields improved resolution at the expense of longer run times, is typically used for DNA sequencing applications. Instrument operation and data collection on the ABI 310 Genetic Analyzer is controlled by a series of steps and procedures that grouped together are referred to as a ‘module’. The standard module used for STR typing is titled ‘GS STR POP 4 (1 m. L) F’ as below. Capillary fill, Pre-electrophoresis, Water wash of capillary, Sample injection, Water wash of capillary, Water dip, Electrophoresis, Detection. This entire process is accomplished in approximately 30 minutes per sample. DNA fragments up to approximately 400 bp in size should be through the capillary within 24 minutes of electrophoresis at 15 000 V on a 47 cm capillary December 15, 2021 CEMB, Lahore 14

December 15, 2021 CEMB, Lahore 15

December 15, 2021 CEMB, Lahore 16

December 15, 2021 CEMB, Lahore 17

December 15, 2021 CEMB, Lahore 18

December 15, 2021 CEMB, Lahore 19

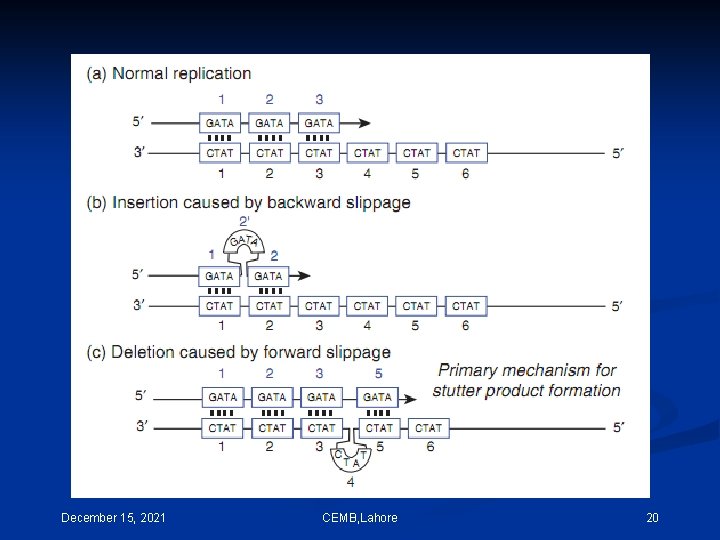
December 15, 2021 CEMB, Lahore 20

December 15, 2021 CEMB, Lahore 21

December 15, 2021 CEMB, Lahore 22

Product rule: If each locus is inherited independent of the other loci, then a calculation of a DNA profile frequency can be made by multiplying each individual genotype frequency together December 15, 2021 CEMB, Lahore 23

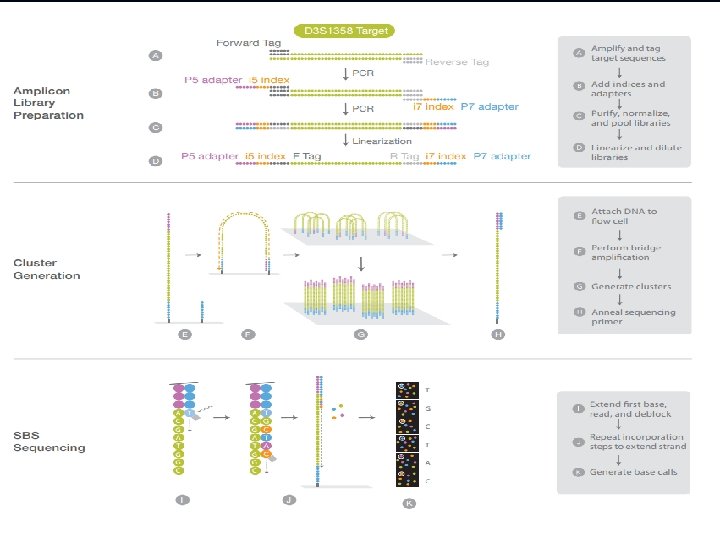
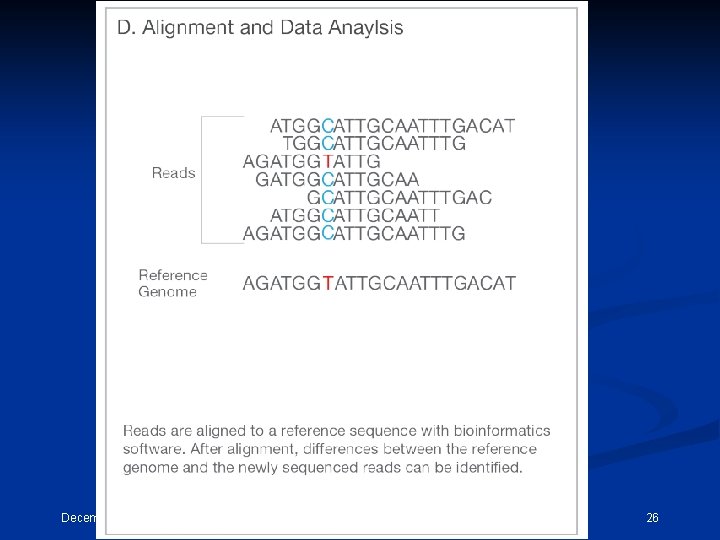
Next Generation Sequencing In principle, the concept behind NGS technology is similar to CE sequencing—DNA polymerase catalyzes the incorporation of fluorescently labeled deoxyribonucleotide triphosphates (d. NTPs) into a DNA template strand during sequential cycles of DNA synthesis. During each cycle, at the point of incorporation, the nucleotides are identified by fluorophore excitation. One critical difference is that, rather than sequencing a single DNA fragment, NGS extends this process across millions of fragments in a massively parallel fashion. December 15, 2021 CEMB, Lahore 24

December 15, 2021 CEMB, Lahore 25

December 15, 2021 CEMB, Lahore 26

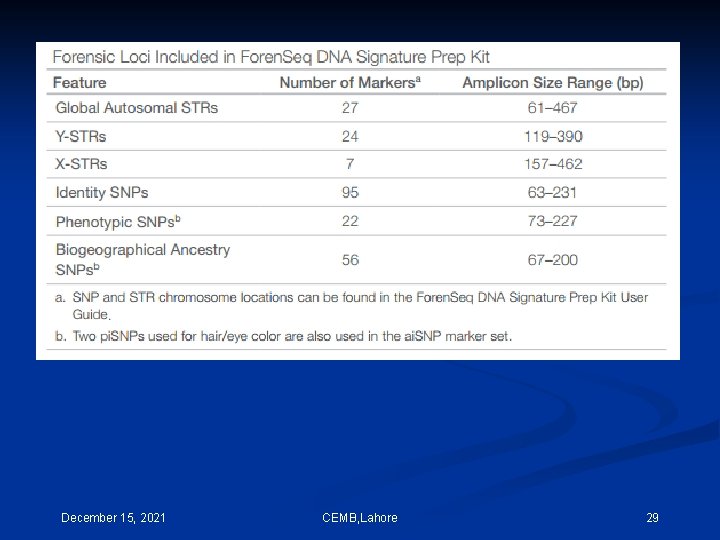
a pioneer study was performed by Zajac and colleagues in 2009, who analyzed three CODIS STR loci, TPOX, CSF 1 PO and D 18 S 51, using the trinucleotide threading (Tn. T) approach by 454 Genome Sequencing System. Subsequently, Irwin et al. in 2011 analyzed 13 CODIS STR loci using the 454 GS Junior system in combination with multiplex identifier technology for single source samples. Bornman et al. in 2012 went further to show that high-throughput sequencing technology can accurately identify the 13 CODIS STR loci as well as the AMEL gene for not only single source but also mixed samples. To process the forensic NGS data, Warshauer et al. in 2013 developed STRait Razor, a software that can analyze the NGS data for 44 STRs, including 23 autosomal and 21 Y chromosome STRs. In addition, Van Neste et al. in 2014 used Illumina’s Mi. Seq system to establish a reference allele database to detect single source and mixed DNA samples; they observed that most locus genotyping results were stable and reliable. December 15, 2021 CEMB, Lahore 27

December 15, 2021 CEMB, Lahore 28

December 15, 2021 CEMB, Lahore 29