Cancer Diagnostic Features Representation in OMOP CDM November

Cancer Diagnostic Features Representation in OMOP CDM November 28, 2017

Considerations • Primary cancer diagnosis – In our current approach, we define cancer diagnosis as a combination of histology (morphology) + topography (anatomy) • Additional diagnostic features – In cancer, features like stage (pathological and clinical), grade, laterality, focality, and some others, are critical to diagnosis differentiation, prognosis, and choice of treatment. These features must accompany primary cancer diagnosis • Association of additional diagnostic features with primary diagnosis – These features are measured when a patient is first diagnosed and also (possibly) for each cancer recurrence – There a possibility of repeated measurements for the same recurrence – There is a possibility of measurements performed/reported on different dates
Modeling and ETL challenges • One pre-coordinated diagnosis concept or multiple diagnosis modifiers? – Pre-coordinated concepts may not work • • Source representation is by axes Certain axes may contain multiple values or variables Too many axes and permutations to maintain Missing axes will prompt using diagnoses of different levels of granularity (if exist) and complicate queries • Temporal association of diagnosis modifiers with primary diagnosis – There should be one set of “verified” cancer modifiers associated with the initial diagnosis (first cancer occurrence) and, possibly, with each recurrence – Repeated measurements of the same modifier (lymph node invasion) may be recorded – Different modifiers may be recorded on different dates • Identification of cancer recurrences (condition era) – First cancer occurrence and further recurrences may be derived algorithmically or extracted from the source data directly. If this information is available in the source, it should be persisted and override the derived one. This mixed approach is new to OMOP – Algorithmic derivation of recurrences would not be the same as current condition era derivation
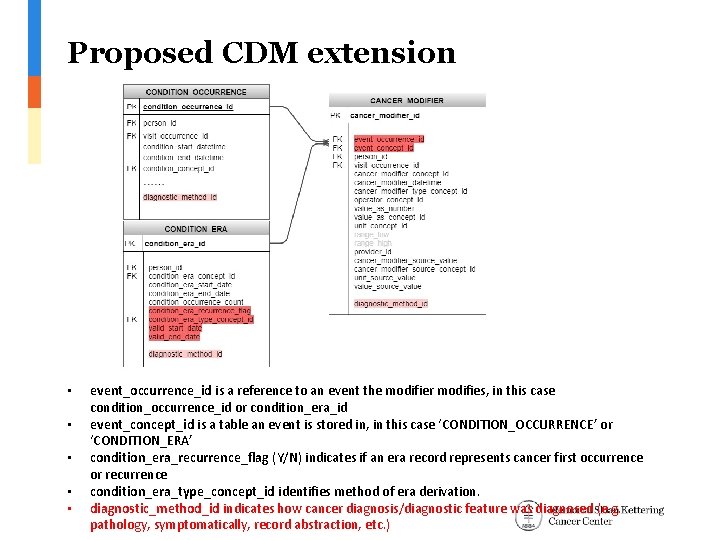
Proposed CDM extension • • • event_occurrence_id is a reference to an event the modifier modifies, in this case condition_occurrence_id or condition_era_id event_concept_id is a table an event is stored in, in this case ‘CONDITION_OCCURRENCE’ or ‘CONDITION_ERA’ condition_era_recurrence_flag (Y/N) indicates if an era record represents cancer first occurrence or recurrence condition_era_type_concept_id identifies method of era derivation. diagnostic_method_id indicates how cancer diagnosis/diagnostic feature was diagnosed (e. g. pathology, symptomatically, record abstraction, etc. )
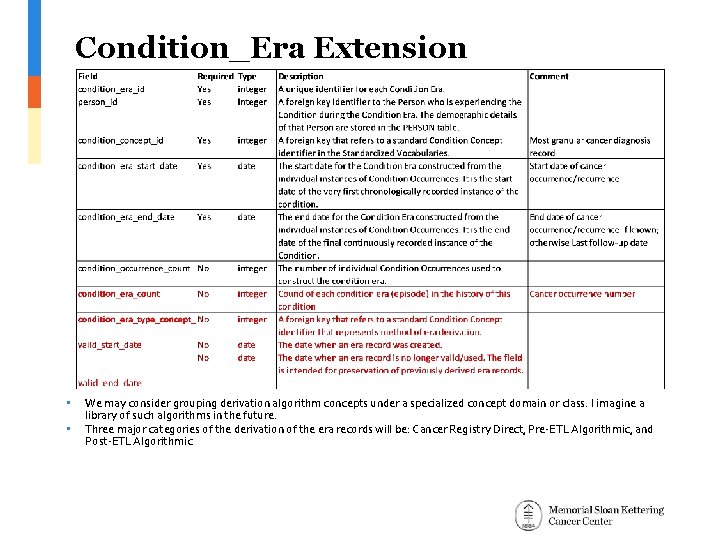
Condition_Era Extension • • We may consider grouping derivation algorithm concepts under a specialized concept domain or class. I imagine a library of such algorithms in the future. Three major categories of the derivation of the era records will be: Cancer Registry Direct, Pre-ETL Algorithmic, and Post-ETL Algorithmic

Alternatives for representing cancer occurrence/recurrences 1. condition_era_recurrence_flag (Y/N) Y indicates a recurrence, N first occurrence – Pros: derivation is not overly complicated – Pros: recurrence/occurrence records are explicitly expressed; no additional processing is required to query them
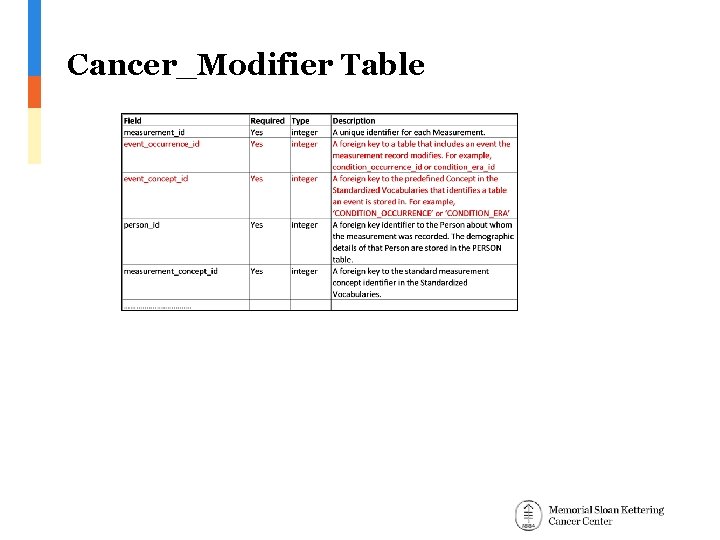
Cancer_Modifier Table
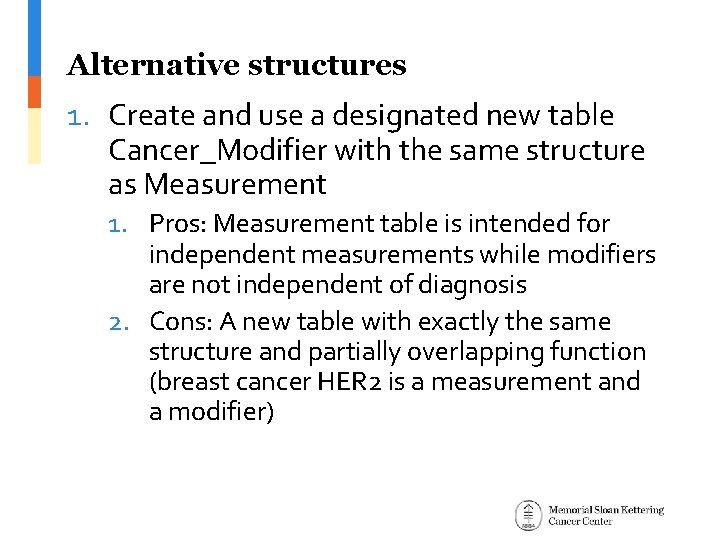
Alternative structures 1. Create and use a designated new table Cancer_Modifier with the same structure as Measurement 1. Pros: Measurement table is intended for independent measurements while modifiers are not independent of diagnosis 2. Cons: A new table with exactly the same structure and partially overlapping function (breast cancer HER 2 is a measurement and a modifier)

Proposed CDM conventions • Diagnosis modifiers are stored in the Cancer_Modifier table • Association of diagnosis modifiers with primary diagnosis – One or multiple condition occurrence records containing primary cancer diagnosis may have associated diagnosis modifiers – Repeated modifier records (lymph node invasion) may be associated with one or multiple condition occurrence records – Modifiers may be recorded on different dates • Representation of cancer recurrences – Cancer recurrences are recorded as condition eras – Each cancer era record is flagged as first occurrence or recurrence – Occurrences/Recurrences are either derived algorithmically or extracted from the source data directly. – Multiple condition eras for the same cancer occurrence/recurrence derived using different methods can be stored. Provenance is a critical attribute of these records. Invalid/unused versions are indicated by valid_end_date. – Algorithmic derivation of recurrences is TBD but will not be the same as current condition era derivation • Association of diagnosis modifiers with cancer recurrences – One set of “verified” cancer modifiers is recorded in Cancer_Modifier and associated with the first cancer occurrence and, if possible, with recurrences
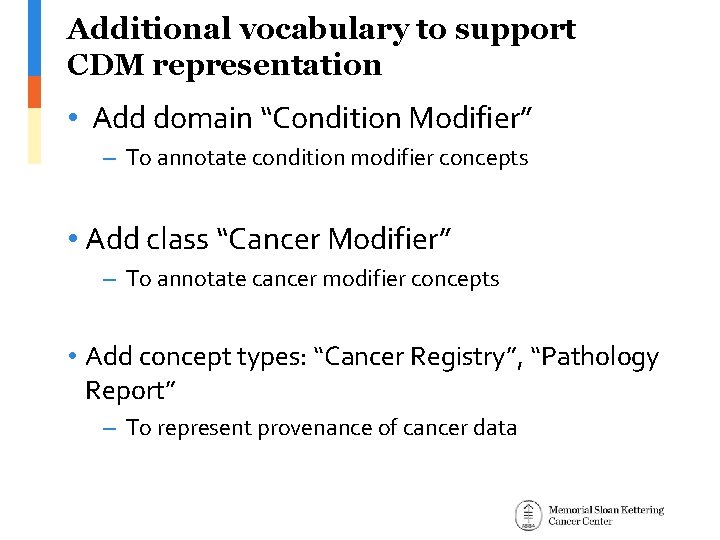
Additional vocabulary to support CDM representation • Add domain “Condition Modifier” – To annotate condition modifier concepts • Add class “Cancer Modifier” – To annotate cancer modifier concepts • Add concept types: “Cancer Registry”, “Pathology Report” – To represent provenance of cancer data

Proposed Vocabulary Approach • Reference Cancer Protocol Templates – – – Issued by CAP (College of American Pathologists) Provide guidelines for collecting the essential data elements for complete reporting of malignant tumors for 88 cancer types Include pathological findings and genomic biomarkers • Use standardized terminology from Nebraska Lexicon Project – – Works under the umbrella of LOINC-SNOMED CT compatible observables harmonization of content between LOINC® and SNOMED CT® Intends to implement CAP Protocol Templates by providing terminology binding between LOINC and SNOMED CT The majority of the associated terminology development is modeled within SNOMED 363787002|Observable entity| hierarchy. Coded LOINC observables are linked to SNOMED value sets. Developed for breast and colorectal cancers. • Implementation in the OMOP Vocabulary – – Adopt Nebraska terminology relationships for implemented cancer types Create OMOP relationships for other cancer types based on CAP Protocol Templates • – Collaborate with UNMS Replace OMOP relationships with Nebraska ones as they become available

CAP Protocol for Invasive Breast Cancer
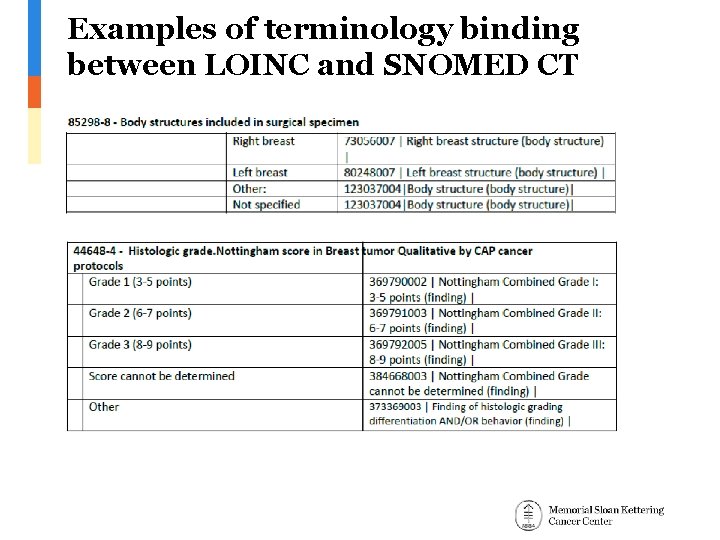
Examples of terminology binding between LOINC and SNOMED CT
Example • Extraction from EMR • Extraction from Cancer Registry
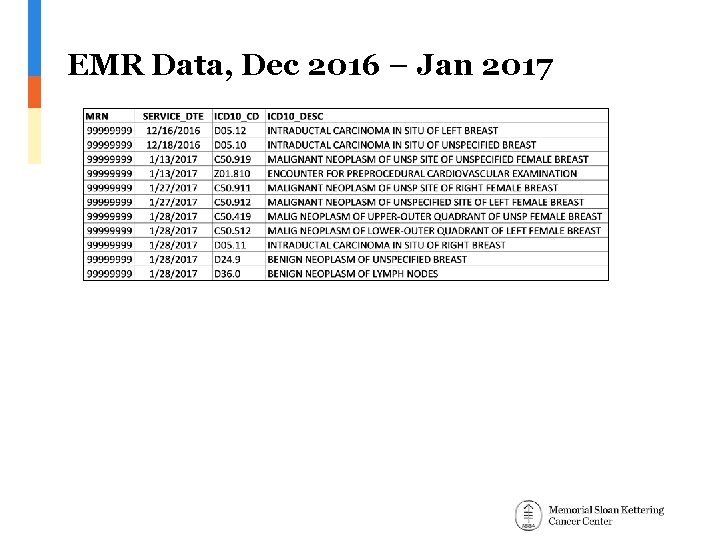
EMR Data, Dec 2016 – Jan 2017
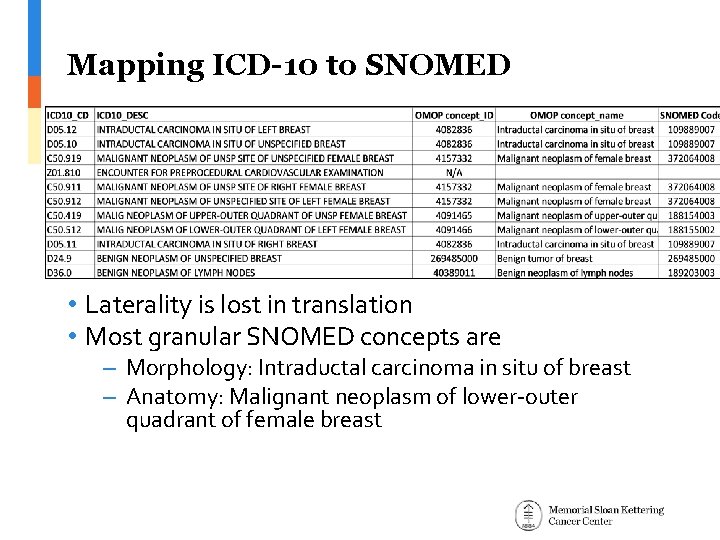
Mapping ICD-10 to SNOMED • Laterality is lost in translation • Most granular SNOMED concepts are – Morphology: Intraductal carcinoma in situ of breast – Anatomy: Malignant neoplasm of lower-outer quadrant of female breast
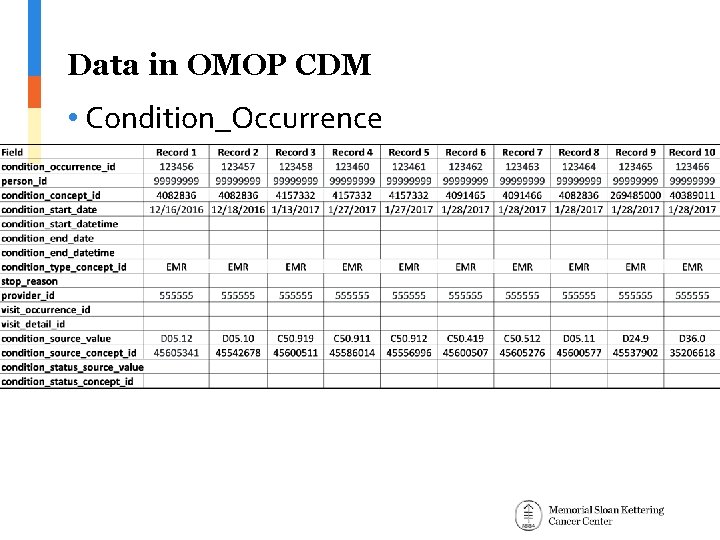
Data in OMOP CDM • Condition_Occurrence
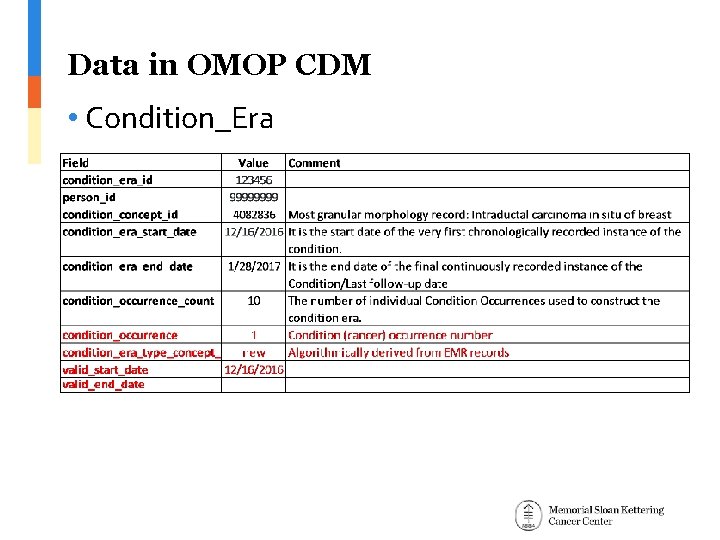
Data in OMOP CDM • Condition_Era
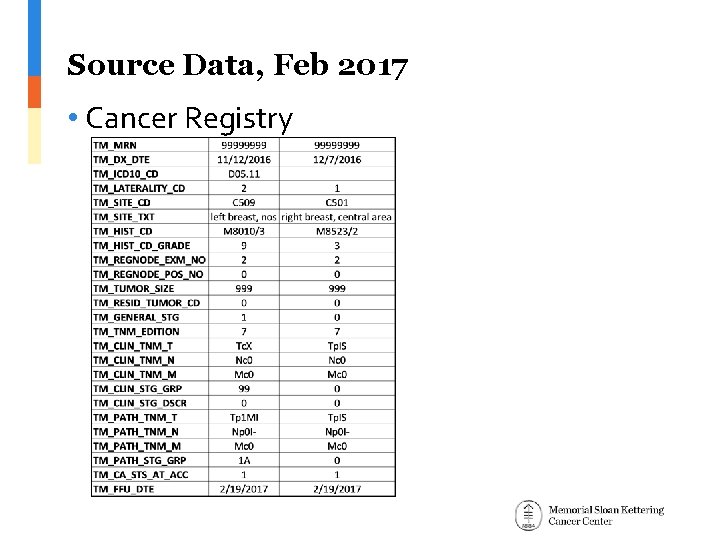
Source Data, Feb 2017 • Cancer Registry
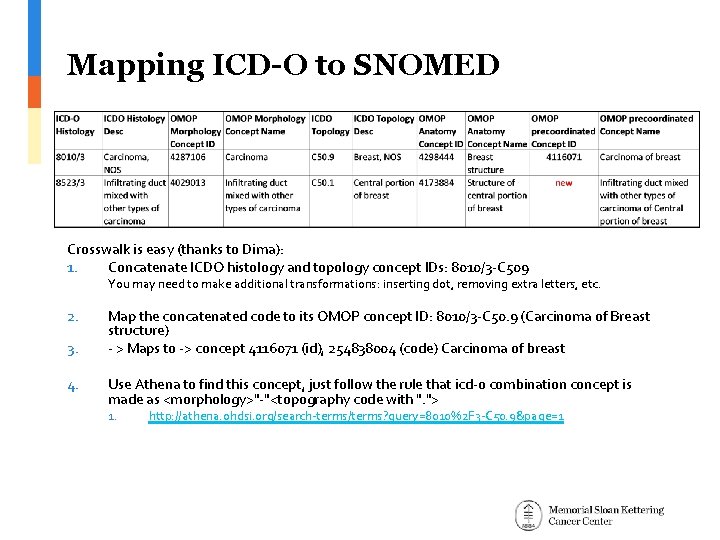
Mapping ICD-O to SNOMED Crosswalk is easy (thanks to Dima): 1. Concatenate ICDO histology and topology concept IDs: 8010/3 -C 509 You may need to make additional transformations: inserting dot, removing extra letters, etc. 2. 3. 4. Map the concatenated code to its OMOP concept ID: 8010/3 -C 50. 9 (Carcinoma of Breast structure) - > Maps to -> concept 4116071 (id), 254838004 (code) Carcinoma of breast Use Athena to find this concept, just follow the rule that icd-o combination concept is made as <morphology>"-"<topography code with ". "> 1. http: //athena. ohdsi. org/search-terms/terms? query=8010%2 F 3 -C 50. 9&page=1
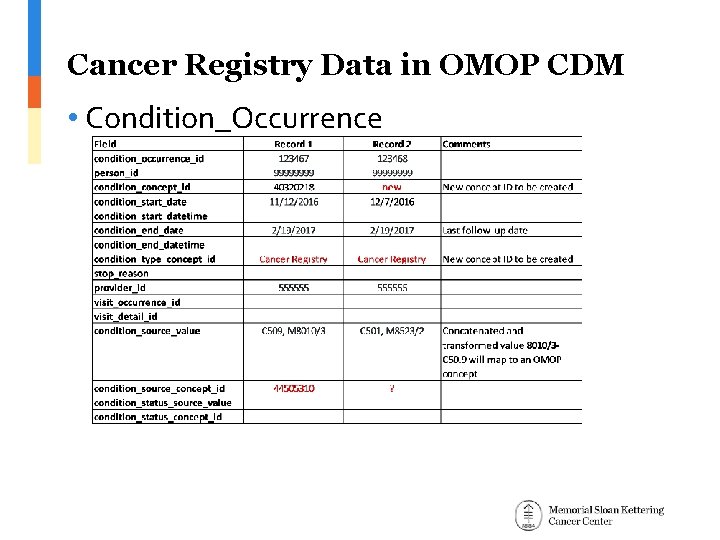
Cancer Registry Data in OMOP CDM • Condition_Occurrence
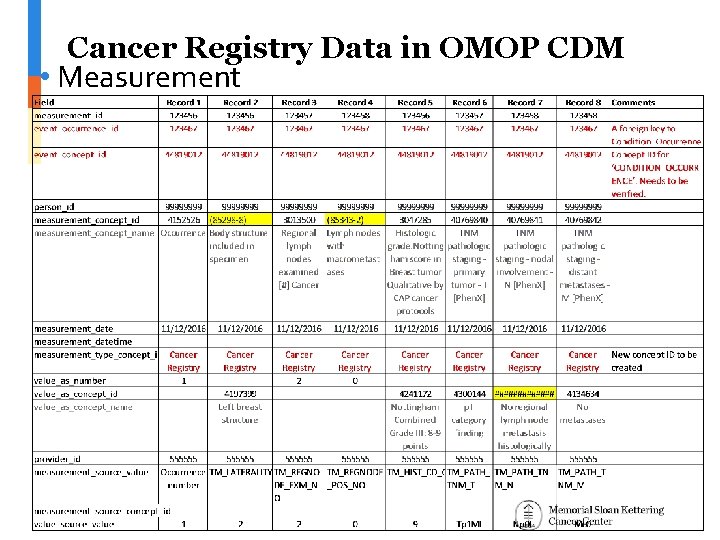
Cancer Registry Data in OMOP CDM • Measurement
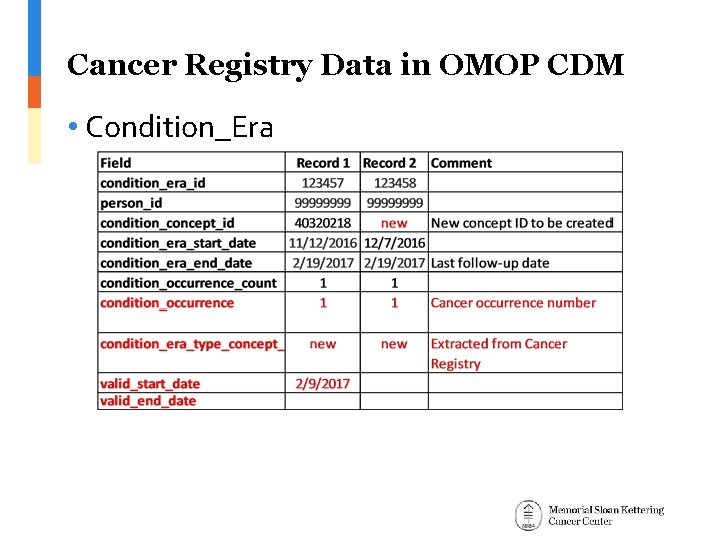
Cancer Registry Data in OMOP CDM • Condition_Era
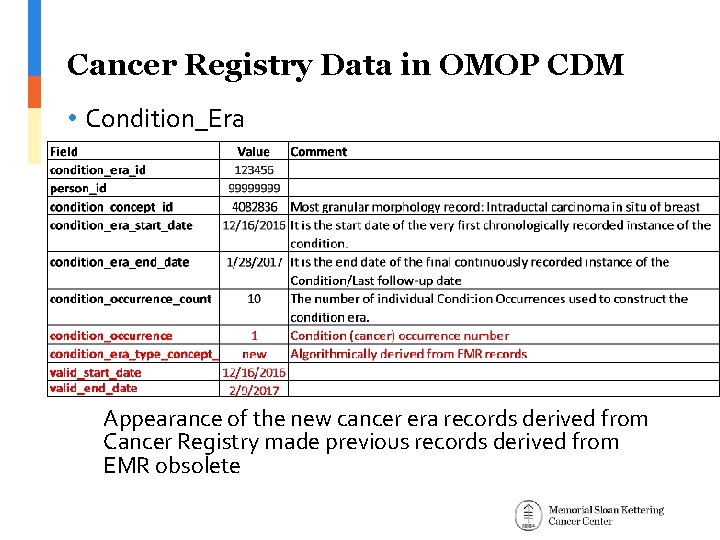
Cancer Registry Data in OMOP CDM • Condition_Era Appearance of the new cancer era records derived from Cancer Registry made previous records derived from EMR obsolete
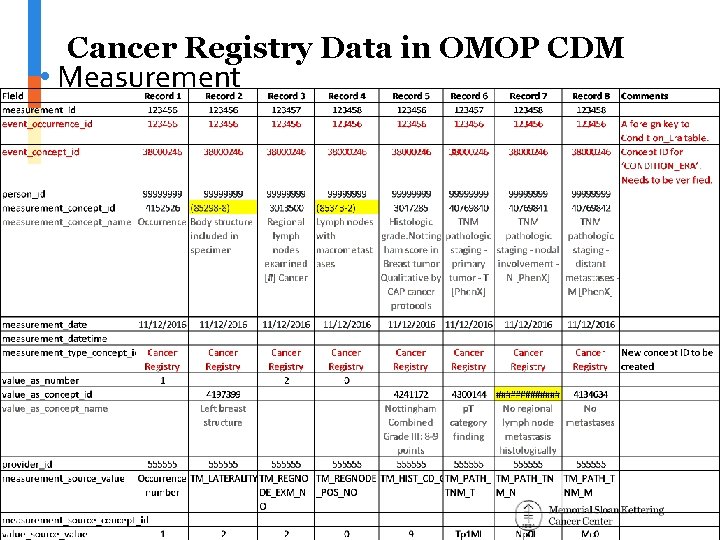
Cancer Registry Data in OMOP CDM • Measurement
References • CAP Protocol Templates http: //www. cap. org/web/oracle/webcenter/portala pp/pagehierarchy/cancer_protocol_templates. jsp x • Nebraska Lexicon https: //www. unmc. edu/pathology/informatics/tdc. html • FORDS https: //www. facs. org/~/media/files/quality%20 prog rams/cancer/ncdb/fords%202016. ashx

Appendix • Astrocytoma turned into GBM – new cancer or recurrence?
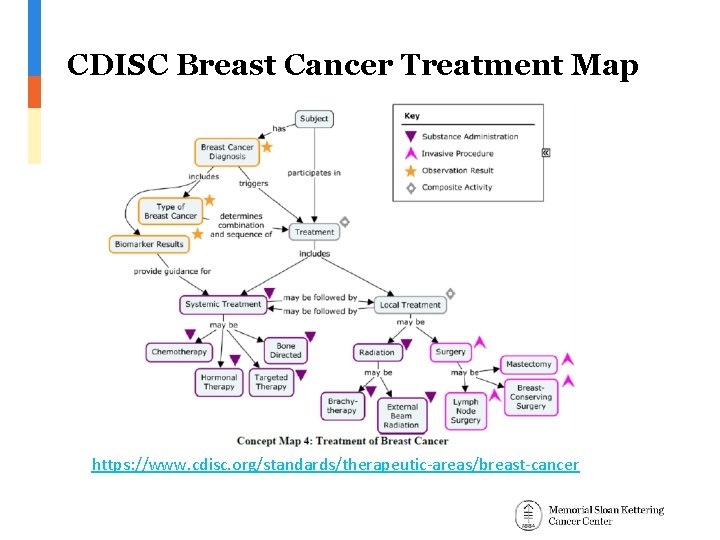
CDISC Breast Cancer Treatment Map https: //www. cdisc. org/standards/therapeutic-areas/breast-cancer

Data Sources • EMR, Structured • Cancer Registry, Structured • Clinical Trials , Structured • Pathology Reports, Unstructured
- Slides: 29