Can Cytogenetics and FISH survive in the modern

![ISCN: 46, XX, t(8; 21)(q 22; q 22)[19]/46, XX[1] ISCN: 46, XX, t(8; 21)(q 22; q 22)[19]/46, XX[1]](https://slidetodoc.com/presentation_image_h/bbbd2546382010d36c6b441ea1499771/image-5.jpg)
![45, X, -Y, t(8; 20)(q 22; p 13), del(11)(q 21 q 25)[18/20] 45, X, -Y, t(8; 20)(q 22; p 13), del(11)(q 21 q 25)[18/20]](https://slidetodoc.com/presentation_image_h/bbbd2546382010d36c6b441ea1499771/image-7.jpg)




- Slides: 48

Can Cytogenetics and FISH survive in the modern genomic era? Application of Cytogenetic, FISH and Microarray Analysis in Diagnosis of Leukemia and Lymphoma Yanming Zhang M. D. Associate Professor, Department of Pathology, Medical Director, Cytogenetics Laboratory, Northwestern University Feinberg School of Medicine

Cytogenetics Laboratory at Northwestern Memorial Hospital, Northwestern University • State-of-the art clinical cytogenetics laboratory with CLIA and CAP certification. • Opened on October 3, 2011, with an average case load of 2000 hematological neoplasms and 150 breast, brain and lung cancer samples (PET FISH). • Staffed with 8 technologists, one resource coordinator, one technical coordinator, one manager and one medical director. • Techniques: Conventional cytogenetic analysis Fluorescence in situ hybridization (FISH) Paraffin embedded tissue (PET)-FISH Genomic SNP microarray

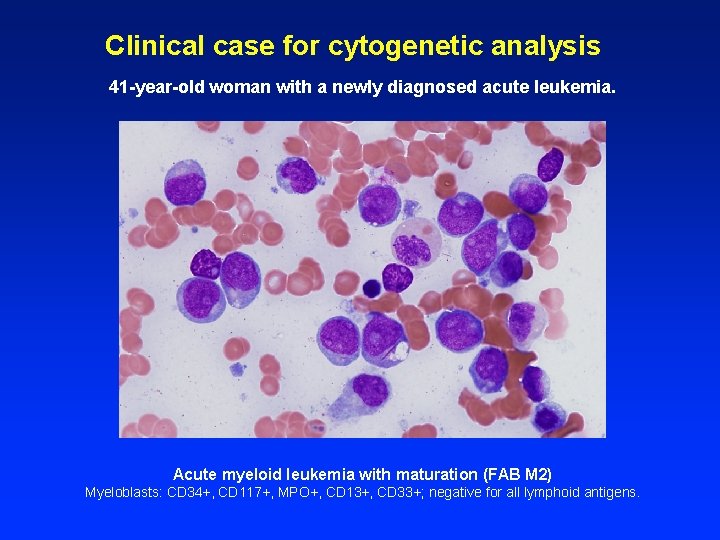
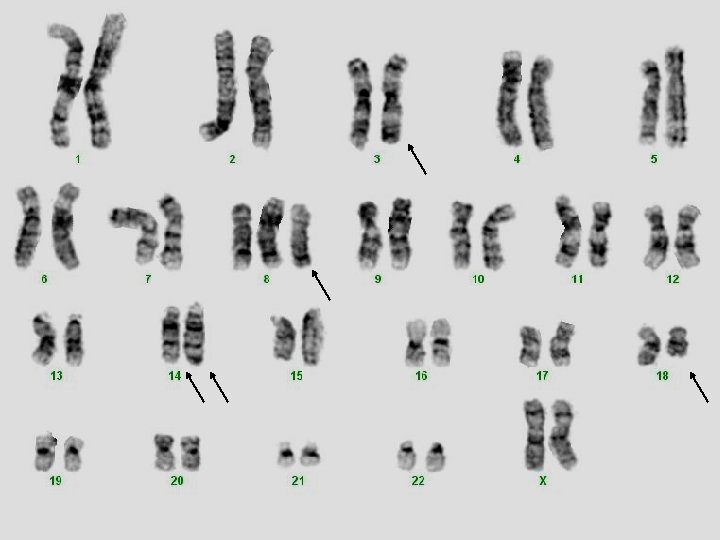
Clinical case for cytogenetic analysis 41 -year-old woman with a newly diagnosed acute leukemia. Acute myeloid leukemia with maturation (FAB M 2) Myeloblasts: CD 34+, CD 117+, MPO+, CD 13+, CD 33+; negative for all lymphoid antigens.

![ISCN 46 XX t8 21q 22 q 221946 XX1 ISCN: 46, XX, t(8; 21)(q 22; q 22)[19]/46, XX[1]](https://slidetodoc.com/presentation_image_h/bbbd2546382010d36c6b441ea1499771/image-5.jpg)
ISCN: 46, XX, t(8; 21)(q 22; q 22)[19]/46, XX[1]

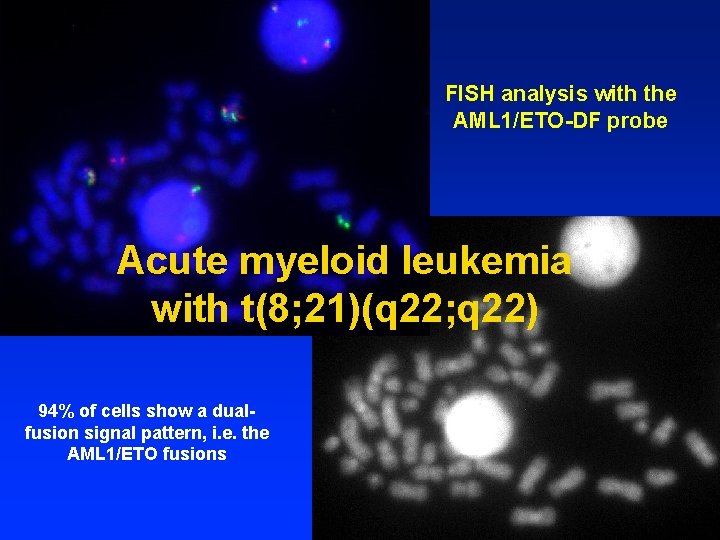
FISH analysis with the AML 1/ETO-DF probe Acute myeloid leukemia with t(8; 21)(q 22; q 22) 94% of cells show a dualfusion signal pattern, i. e. the AML 1/ETO fusions
![45 X Y t8 20q 22 p 13 del11q 21 q 251820 45, X, -Y, t(8; 20)(q 22; p 13), del(11)(q 21 q 25)[18/20]](https://slidetodoc.com/presentation_image_h/bbbd2546382010d36c6b441ea1499771/image-7.jpg)
45, X, -Y, t(8; 20)(q 22; p 13), del(11)(q 21 q 25)[18/20]

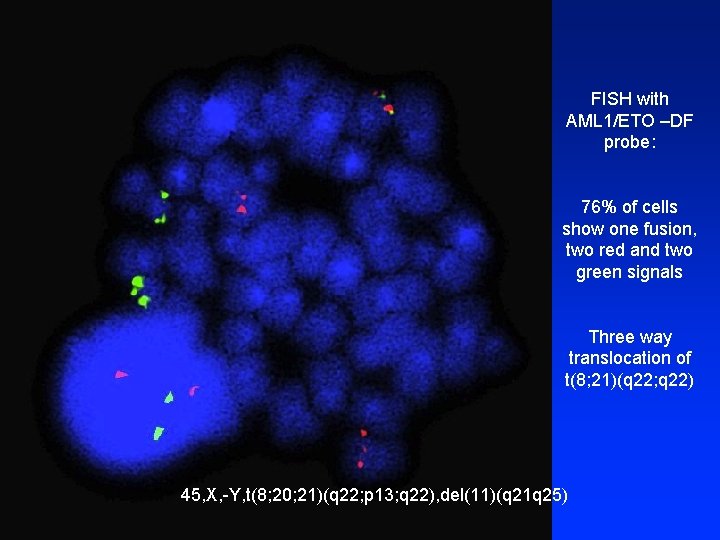
FISH with AML 1/ETO –DF probe: 76% of cells show one fusion, two red and two green signals Three way translocation of t(8; 21)(q 22; q 22) 45, X, -Y, t(8; 20; 21)(q 22; p 13; q 22), del(11)(q 21 q 25)

Procedure of cytogenetic analysis

Cancer Cytogenetics • Samples: (fresh!) bone marrow (aspirate or core) peripheral blood lymph node/spleen/tonsil solid tumor mass CNS, plural fluids, etc • Culturing: no mitogens added short term cultures (24 hr, 48 hr) • Chromosomes: Leukemia cells with poor morphology and few short and fuzzy bands, whereas normal cells nice bands. • Analysis: Heterogeneous populations (normal, abnormal clones). Precise hematopathological diagnosis is important for targeted detection of recurring chromosome abnormalities in specific subtypes.

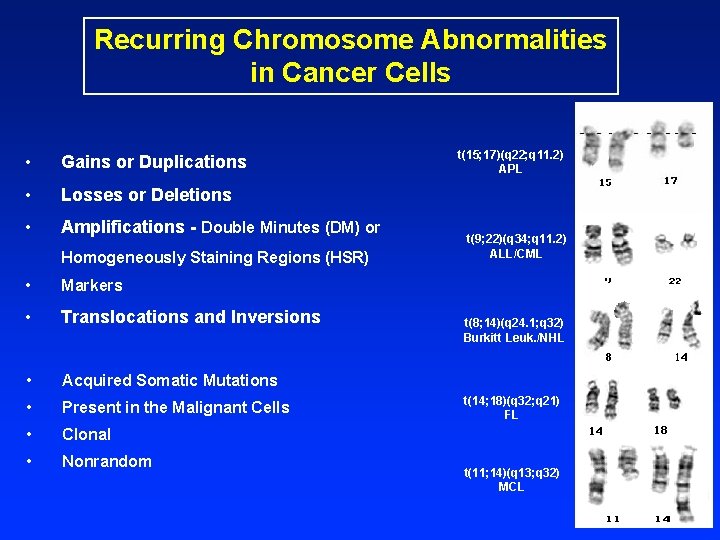
Recurring Chromosome Abnormalities in Cancer Cells • Gains or Duplications • Losses or Deletions • Amplifications - Double Minutes (DM) or Homogeneously Staining Regions (HSR) • Markers • Translocations and Inversions • Acquired Somatic Mutations • Present in the Malignant Cells • Clonal • Nonrandom t(15; 17)(q 22; q 11. 2) APL t(9; 22)(q 34; q 11. 2) ALL/CML t(8; 14)(q 24. 1; q 32) Burkitt Leuk. /NHL t(14; 18)(q 32; q 21) FL t(11; 14)(q 13; q 32) MCL

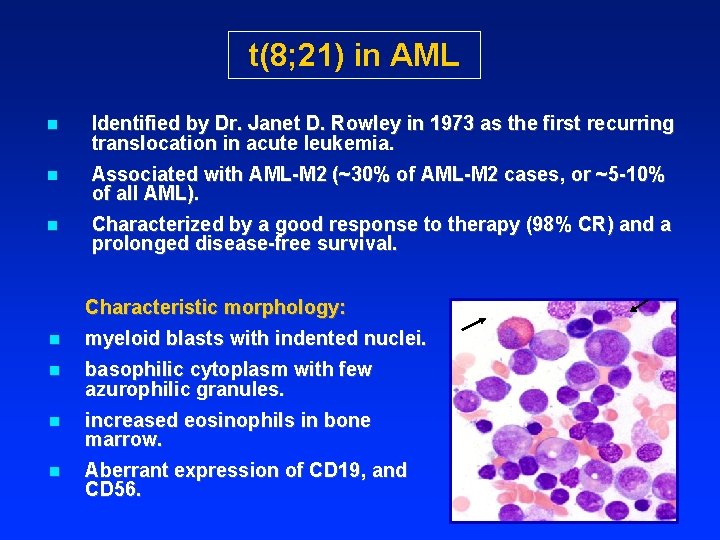
t(8; 21) in AML n Identified by Dr. Janet D. Rowley in 1973 as the first recurring translocation in acute leukemia. n Associated with AML-M 2 (~30% of AML-M 2 cases, or ~5 -10% of all AML). n Characterized by a good response to therapy (98% CR) and a prolonged disease-free survival. Characteristic morphology: n myeloid blasts with indented nuclei. n basophilic cytoplasm with few azurophilic granules. n increased eosinophils in bone marrow. n Aberrant expression of CD 19, and CD 56.

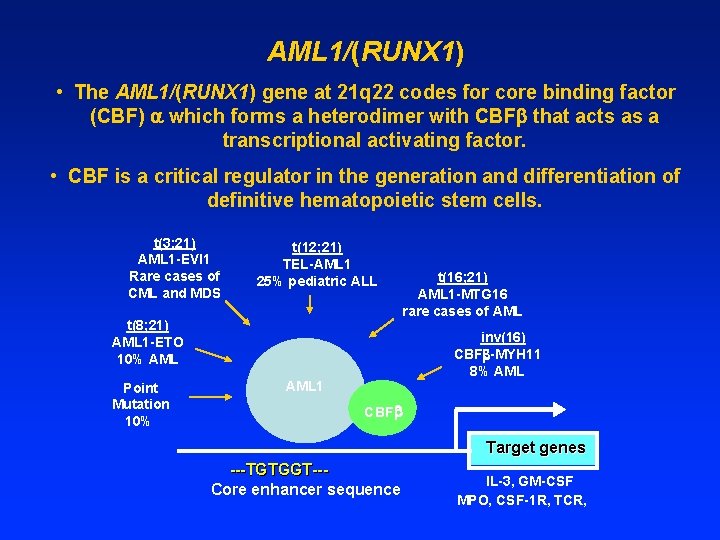
AML 1/(RUNX 1) • The AML 1/(RUNX 1) gene at 21 q 22 codes for core binding factor (CBF) which forms a heterodimer with CBF that acts as a transcriptional activating factor. • CBF is a critical regulator in the generation and differentiation of definitive hematopoietic stem cells. t(3; 21) AML 1 -EVI 1 Rare cases of CML and MDS t(12; 21) TEL-AML 1 25% pediatric ALL t(8; 21) AML 1 -ETO 10% AML Point Mutation 10% t(16; 21) AML 1 -MTG 16 rare cases of AML inv(16) CBF -MYH 11 8% AML 1 CBF Target genes ---TGTGGT--Core enhancer sequence IL-3, GM-CSF MPO, CSF-1 R, TCR,

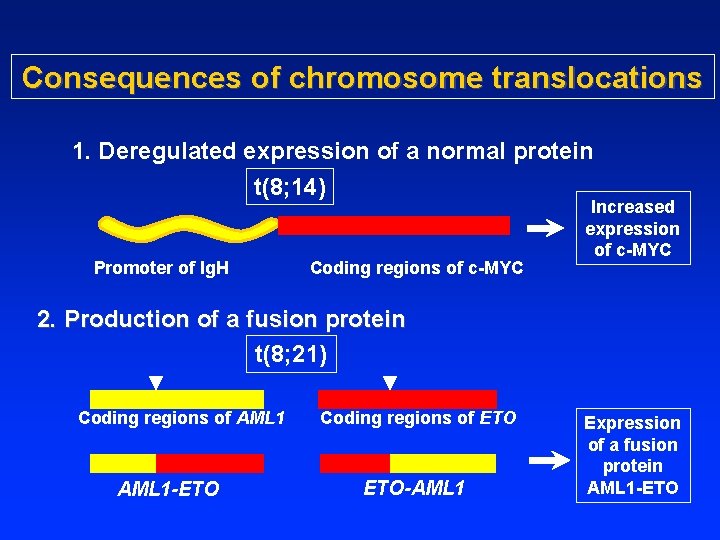
Consequences of chromosome translocations 1. Deregulated expression of a normal protein t(8; 14) Promoter of Ig. H Coding regions of c-MYC Increased expression of c-MYC 2. Production of a fusion protein t(8; 21) Coding regions of AML 1 Coding regions of ETO AML 1 -ETO ETO-AML 1 Expression of a fusion protein AML 1 -ETO

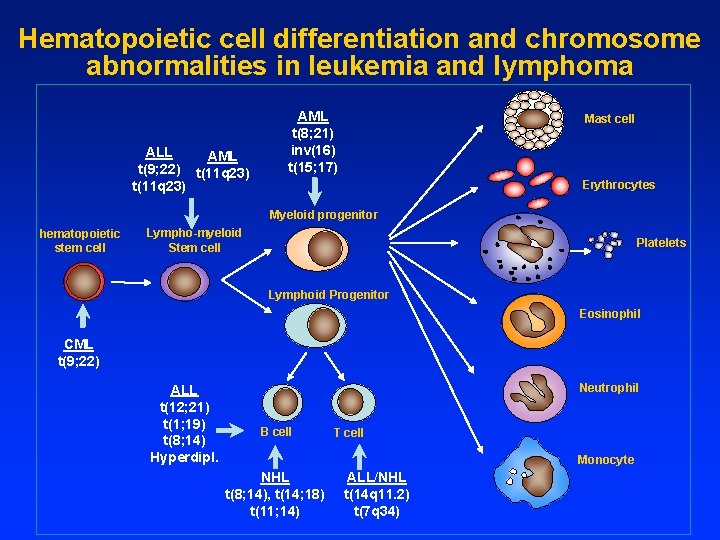
Hematopoietic cell differentiation and chromosome abnormalities in leukemia and lymphoma ALL AML t(9; 22) t(11 q 23) AML t(8; 21) inv(16) t(15; 17) Mast cell Erythrocytes Myeloid progenitor hematopoietic stem cell Lympho-myeloid Stem cell Platelets Lymphoid Progenitor Eosinophil CML t(9; 22) ALL t(12; 21) t(1; 19) t(8; 14) Hyperdipl. Neutrophil B cell T cell Monocyte NHL t(8; 14), t(14; 18) t(11; 14) ALL/NHL t(14 q 11. 2) t(7 q 34)

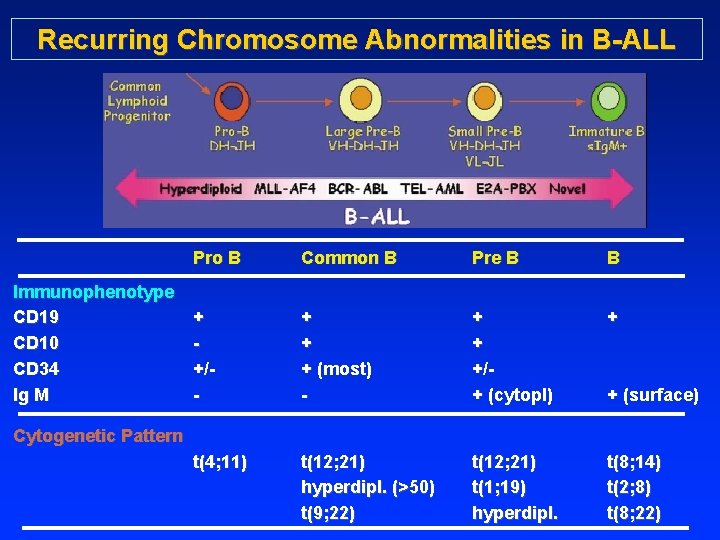
Recurring Chromosome Abnormalities in B-ALL Immunophenotype CD 19 CD 10 CD 34 Ig M Pro B Common B Pre B B + +/- + + + (most) - + + +/+ (cytopl) + t(4; 11) t(12; 21) hyperdipl. (>50) t(9; 22) t(12; 21) t(1; 19) hyperdipl. t(8; 14) t(2; 8) t(8; 22) + (surface) Cytogenetic Pattern

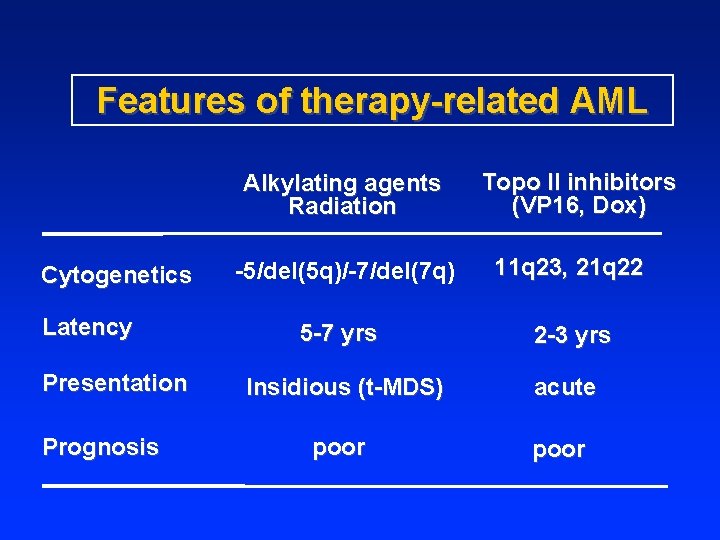
Features of therapy-related AML Alkylating agents Radiation Cytogenetics Latency Presentation Prognosis Topo II inhibitors (VP 16, Dox) -5/del(5 q)/-7/del(7 q) 11 q 23, 21 q 22 5 -7 yrs 2 -3 yrs Insidious (t-MDS) acute poor


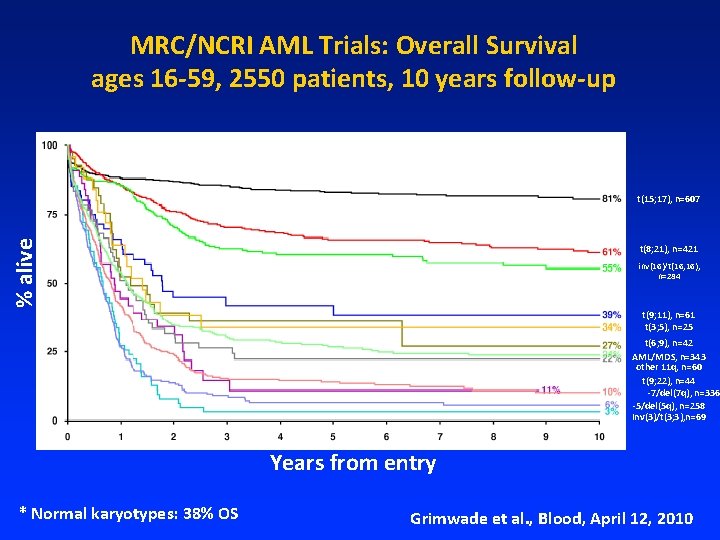
MRC/NCRI AML Trials: Overall Survival ages 16 -59, 2550 patients, 10 years follow-up % alive t(15; 17), n=607 t(8; 21), n=421 inv(16)/t(16; 16), n=284 t(9; 11), n=61 t(3; 5), n=25 t(6; 9), n=42 AML/MDS, n=343 other 11 q, n=60 t(9; 22), n=44 -7/del(7 q), n=336 -5/del(5 q), n=258 Inv(3)/t(3; 3), n=69 Years from entry * Normal karyotypes: 38% OS Grimwade et al. , Blood, April 12, 2010

Clinical significance of chromosome abnormalities in leukemia and lymphoma • Diagnosis and differential diagnosis: WHO classification based on specific cytogenetic/molecular genetic findings, such as t(8; 21), t(15; 17), inv(16), t(9; 11) and other 11 q 23/MLL, inv(3)/t(3; 3), t(6; 9), t(1; 22). • Treatment protocols: APL: PML/RARa: ATRA+CT. CBF [t(8; 21) and inv(16)]: HDAC consolidation. • Monitoring response and engraftment of BMT cytogenetic complete remission (CR) and MRD • Prognosis: most critical and independent indicators. favorable (55 -81% cured): t(15; 17), inv(16), t(8; 21); intermediate (40%): t(9; 11), normal karyotype; unfavorable (<5%): complex, abnl 5 and 7, inv(3), t(6; 9)

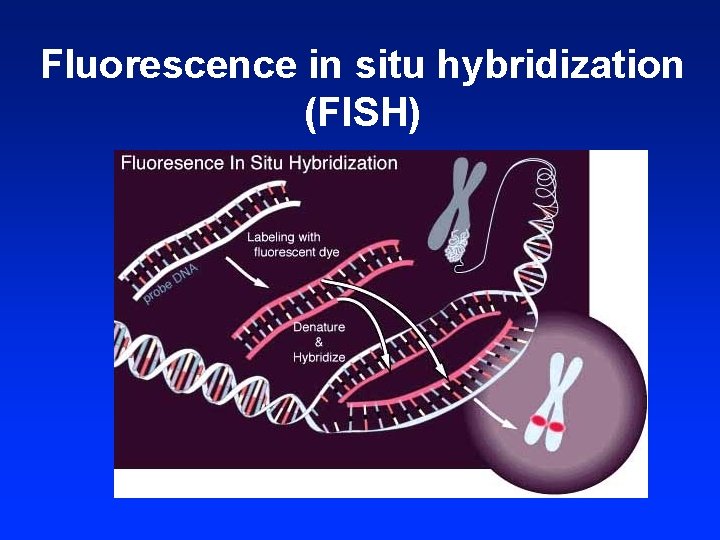
Fluorescence in situ hybridization (FISH)

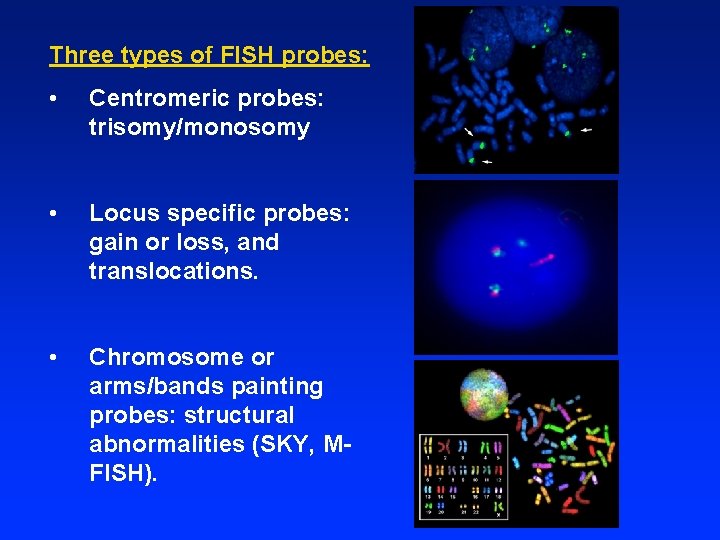
Three types of FISH probes: • Centromeric probes: trisomy/monosomy • Locus specific probes: gain or loss, and translocations. • Chromosome or arms/bands painting probes: structural abnormalities (SKY, MFISH).

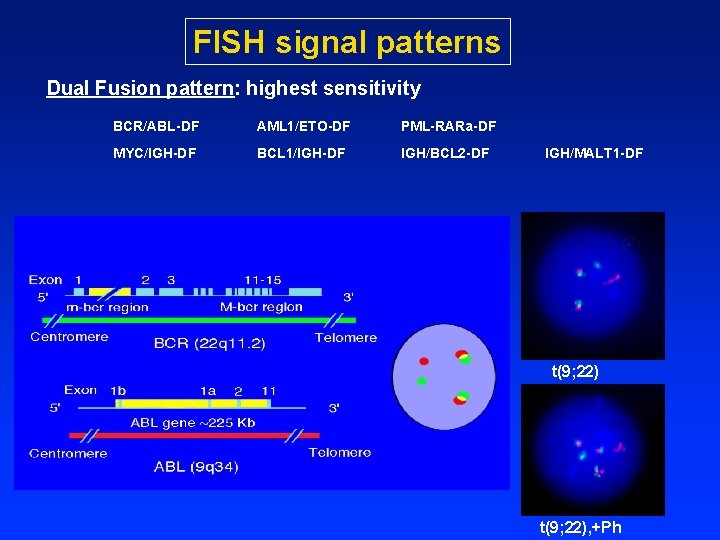
FISH signal patterns Dual Fusion pattern: highest sensitivity BCR/ABL-DF AML 1/ETO-DF PML-RARa-DF MYC/IGH-DF BCL 1/IGH-DF IGH/BCL 2 -DF IGH/MALT 1 -DF t(9; 22), +Ph

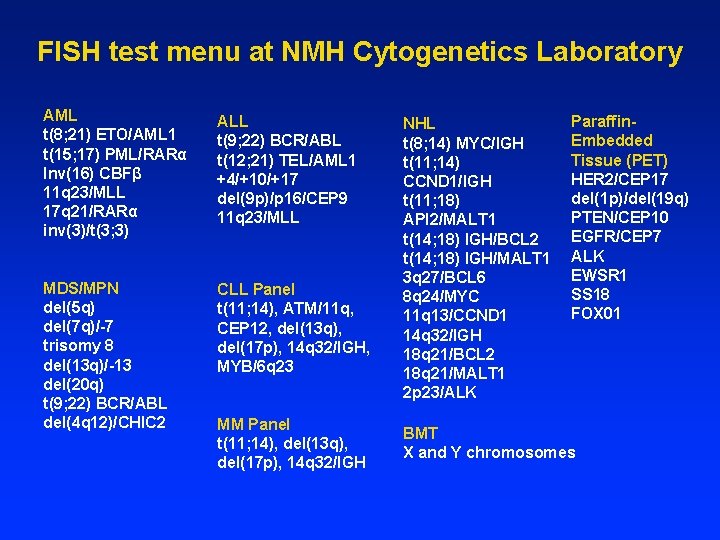
FISH test menu at NMH Cytogenetics Laboratory AML t(8; 21) ETO/AML 1 t(15; 17) PML/RARα Inv(16) CBFβ 11 q 23/MLL 17 q 21/RARα inv(3)/t(3; 3) ALL t(9; 22) BCR/ABL t(12; 21) TEL/AML 1 +4/+10/+17 del(9 p)/p 16/CEP 9 11 q 23/MLL MDS/MPN del(5 q) del(7 q)/-7 trisomy 8 del(13 q)/-13 del(20 q) t(9; 22) BCR/ABL del(4 q 12)/CHIC 2 CLL Panel t(11; 14), ATM/11 q, CEP 12, del(13 q), del(17 p), 14 q 32/IGH, MYB/6 q 23 MM Panel t(11; 14), del(13 q), del(17 p), 14 q 32/IGH NHL t(8; 14) MYC/IGH t(11; 14) CCND 1/IGH t(11; 18) API 2/MALT 1 t(14; 18) IGH/BCL 2 t(14; 18) IGH/MALT 1 3 q 27/BCL 6 8 q 24/MYC 11 q 13/CCND 1 14 q 32/IGH 18 q 21/BCL 2 18 q 21/MALT 1 2 p 23/ALK Paraffin. Embedded Tissue (PET) HER 2/CEP 17 del(1 p)/del(19 q) PTEN/CEP 10 EGFR/CEP 7 ALK EWSR 1 SS 18 FOX 01 BMT X and Y chromosomes

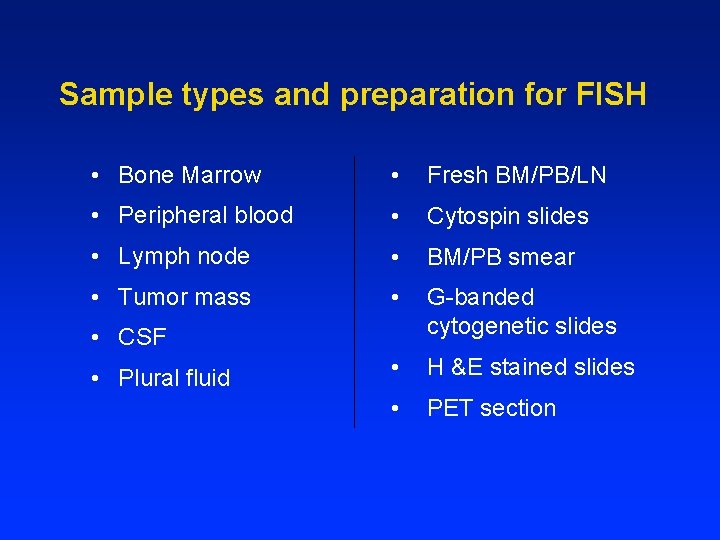
Sample types and preparation for FISH • Bone Marrow • Fresh BM/PB/LN • Peripheral blood • Cytospin slides • Lymph node • BM/PB smear • Tumor mass • G-banded cytogenetic slides • H &E stained slides • PET section • CSF • Plural fluid

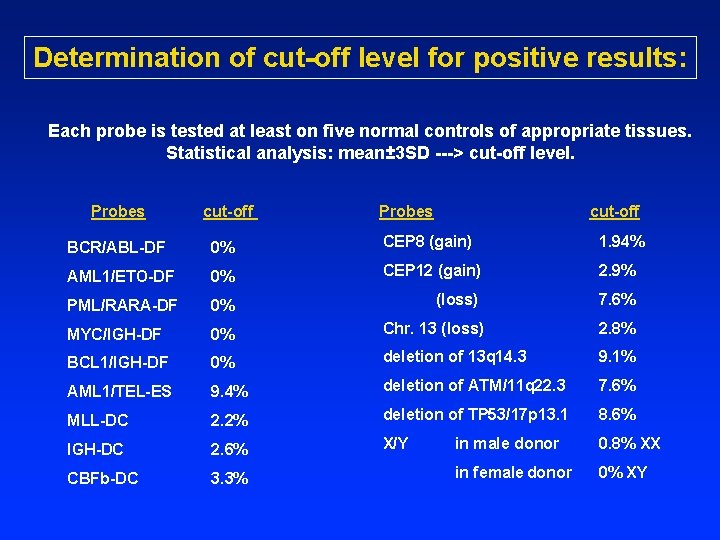
FISH quality control • Each new probe/lot is evaluated with positive and negative controls to assay sensitivity and specificity and to determine the cut-off level. • Negative and positive controls are tested with each probe hybridization with patient samples. • At least two independent observers score for each assay (200 cells per observer).

Determination of cut-off level for positive results: Each probe is tested at least on five normal controls of appropriate tissues. Statistical analysis: mean± 3 SD ---> cut-off level. Probes cut-off Probes cut-off BCR/ABL-DF 0% CEP 8 (gain) 1. 94% AML 1/ETO-DF 0% CEP 12 (gain) 2. 9% PML/RARA-DF 0% (loss) 7. 6% MYC/IGH-DF 0% Chr. 13 (loss) 2. 8% BCL 1/IGH-DF 0% deletion of 13 q 14. 3 9. 1% AML 1/TEL-ES 9. 4% deletion of ATM/11 q 22. 3 7. 6% MLL-DC 2. 2% deletion of TP 53/17 p 13. 1 8. 6% IGH-DC 2. 6% X/Y in male donor 0. 8% XX CBFb-DC 3. 3% in female donor 0% XY


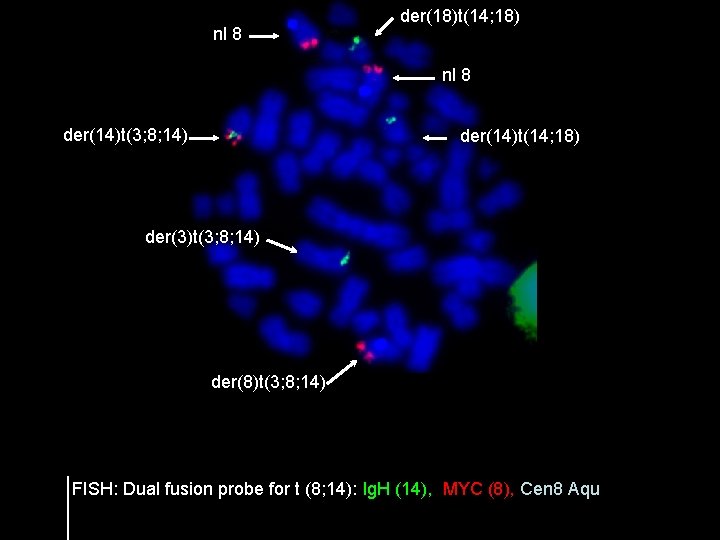
nl 8 der(18)t(14; 18) nl 8 der(14)t(3; 8; 14) der(14)t(14; 18) der(3)t(3; 8; 14) der(8)t(3; 8; 14) FISH: Dual fusion probe for t (8; 14): Ig. H (14), MYC (8), Cen 8 Aqu

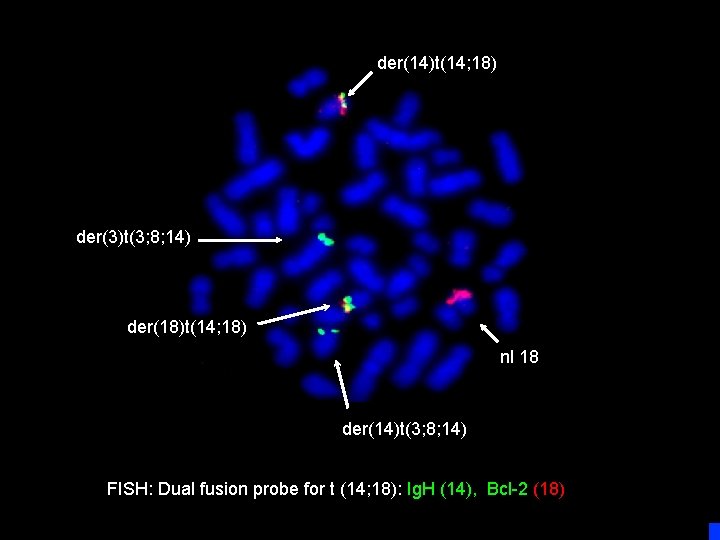
der(14)t(14; 18) der(3)t(3; 8; 14) der(18)t(14; 18) nl 18 der(14)t(3; 8; 14) FISH: Dual fusion probe for t (14; 18): Ig. H (14), Bcl-2 (18)

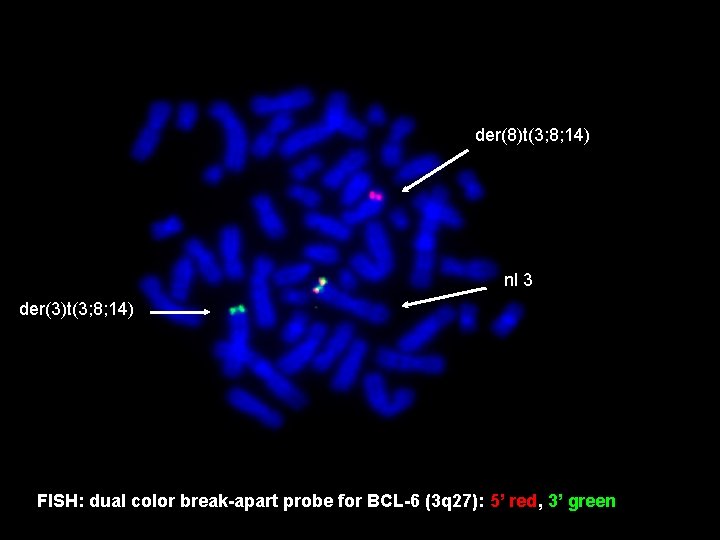
der(8)t(3; 8; 14) nl 3 der(3)t(3; 8; 14) FISH: dual color break-apart probe for BCL-6 (3 q 27): 5’ red, 3’ green

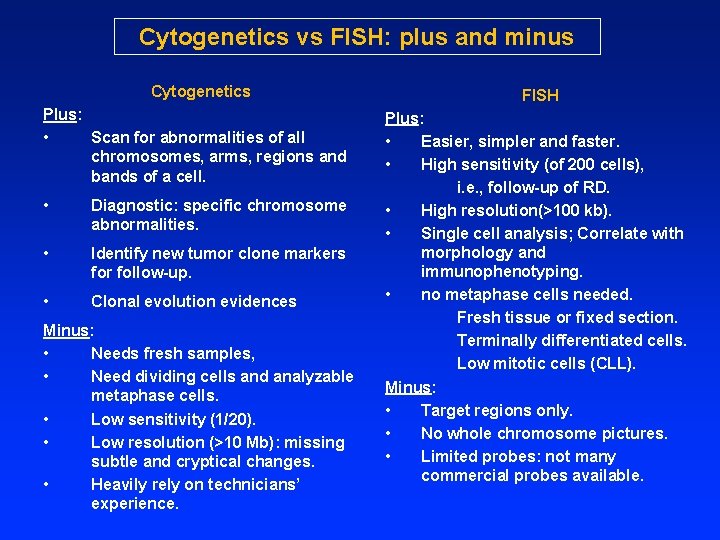
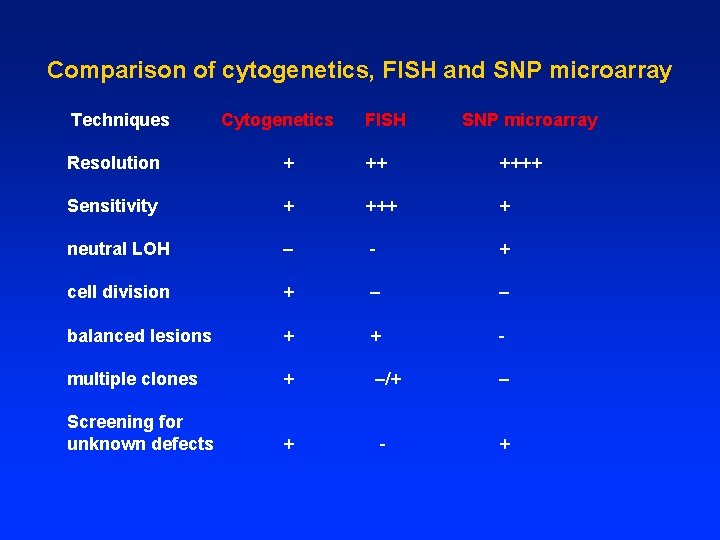
Cytogenetics vs FISH: plus and minus Cytogenetics Plus: • Scan for abnormalities of all chromosomes, arms, regions and bands of a cell. • Diagnostic: specific chromosome abnormalities. • Identify new tumor clone markers for follow-up. • Clonal evolution evidences Minus: • Needs fresh samples, • Need dividing cells and analyzable metaphase cells. • Low sensitivity (1/20). • Low resolution (>10 Mb): missing subtle and cryptical changes. • Heavily rely on technicians’ experience. FISH Plus: • Easier, simpler and faster. • High sensitivity (of 200 cells), i. e. , follow-up of RD. • High resolution(>100 kb). • Single cell analysis; Correlate with morphology and immunophenotyping. • no metaphase cells needed. Fresh tissue or fixed section. Terminally differentiated cells. Low mitotic cells (CLL). Minus: • Target regions only. • No whole chromosome pictures. • Limited probes: not many commercial probes available.

Triaging cytogenetic/FISH analysis indicated • All diagnostic samples of leukemia and lymphoma (confirmed or suspicious). • • • All evolving, transforming or relapsed samples. • 1 st sample after BMT for disease markers or polymorphisms. Residual disease samples if diagnostic samples are not analyzed. All follow-up samples at RD or CR if the diagnostic sample was abnormal in cytogenetic analysis. NOT indicated • • All reactive or benign samples. BM or PB with no involvement of NHL. RD and CR samples if the diagnostic sample was normal (unless there are changes in morphology/immunology). Post-transplant samples with 100% donor cells (XX/XY) or remission sample with no known chromosome abnormalities in FISH study.

Cytogenetics or FISH, or Both tests? • All newly diagnosed AML/MDS cases need cytogenetics first: If specific chromosome abnormalities are known for certain subtype, and cytogenetic analysis is normal, FISH should be added. if rush, FISH for specific chromosome abnormalities may be requested first. • In RD cases with known chromosome abnormalities, such as t(9; 22) in CML, either cytogenetics or FISH are needed. If cytogenetics is negative or inadequate, FISH will be helpful. If cytogenetics is positive, FISH will not provide more information. • At CR or MRD status, FISH is more helpful than cytogenetics in detection of the known chromosome abnormalities (if probes available).

Is ordering a FISH panel for AML, MDS, and NHL justified? NO • Multiple comparison of conventional cytogenetic and FISH tests in several large series of AML and MDS in 1990 s showed that additional common chromosome abnormalities is 2 -4% by FISH using 7 -8 probes in AML and MDS with complete (20 cells) cytogenetic analysis. • FISH panel can detected common chromosome abnormalities in about 30% of AML and MDS with inadequate cytogenetic analysis. Recommendations • Cytogenetic analysis first in all newly diagnosed AML, MDS and MPN. • If cytogenetics is inadequate, FISH with panel is warranted in AML/MDS. • Once a chromosome abnormality is identified at DX, FISH is performed to follow up for disease status and treatment response. • FISH selectively detect recurring translocations in various subtypes of NHL.

Application of genomic array analysis in leukemia and lymphoma ---potentials and problems

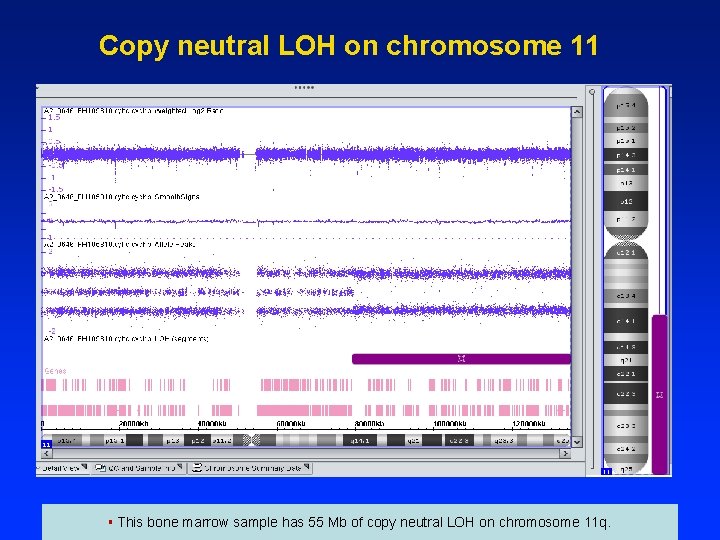
Copy neutral LOH on chromosome 11 § This bone marrow sample has 55 Mb of copy neutral LOH on chromosome 11 q.

Discoveries: Genome-Wide Copy Number Analyses Mullighan et al. Science 2008 322: 1377

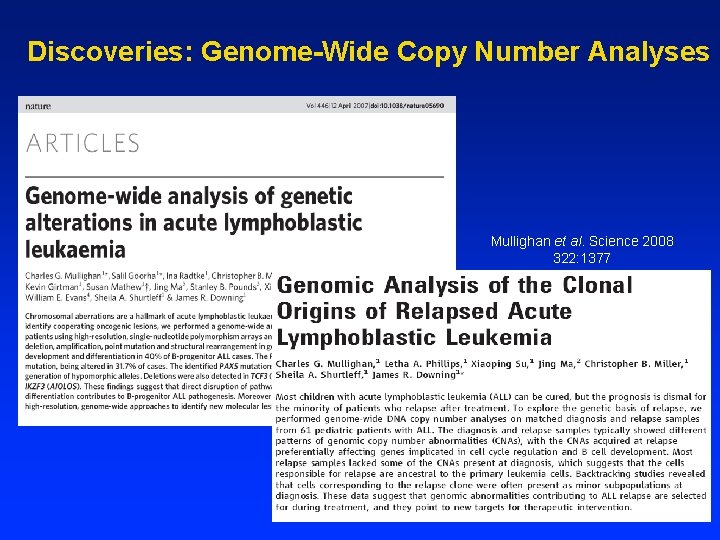
Genome-wide analysis of copy number changes in diagnosis ALL samples

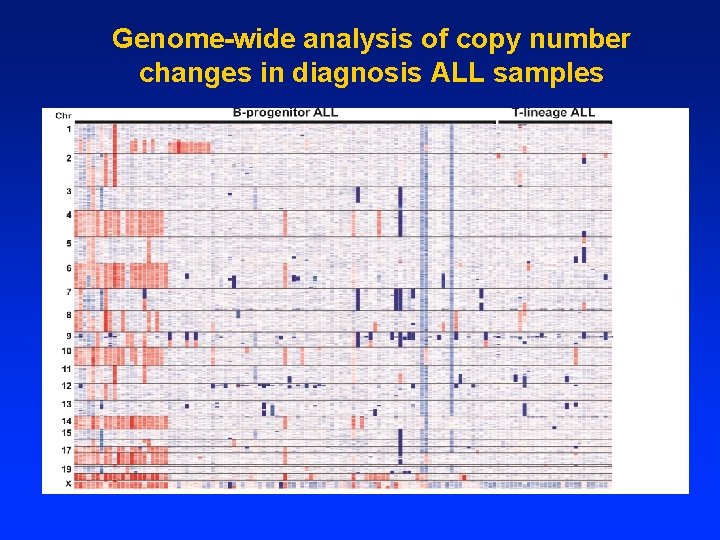
Common clonal origin of relapse and diagnosis samples 1. 89% retained ≥ 1 diagnosis CNA at relapse 2. 86% of pairs shared identical antigen receptor CNA at diagnosis and relapse antigen receptors at diagnosis and relapse

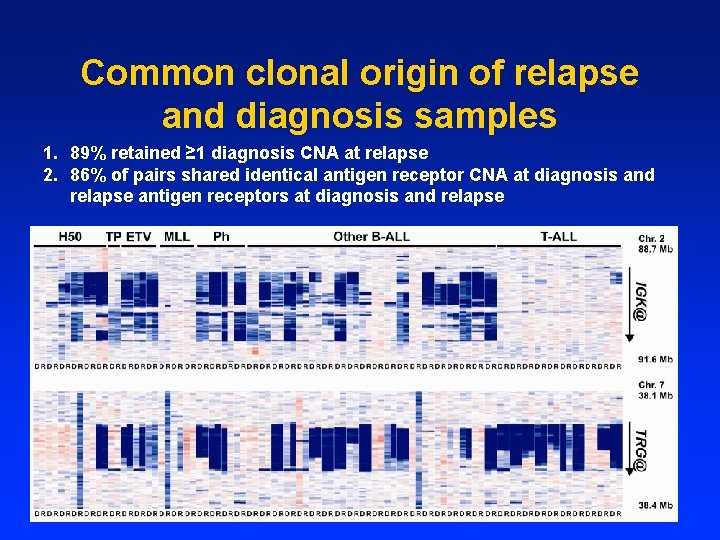
Backtracking relapse-acquired CNA • Clonal evolution, or relapse clone present at low levels at diagnosis? • PCR assays for 10 relapse-acquired CNA: 7 present at diagnosis

Evolution of diagnosis and relapse clones

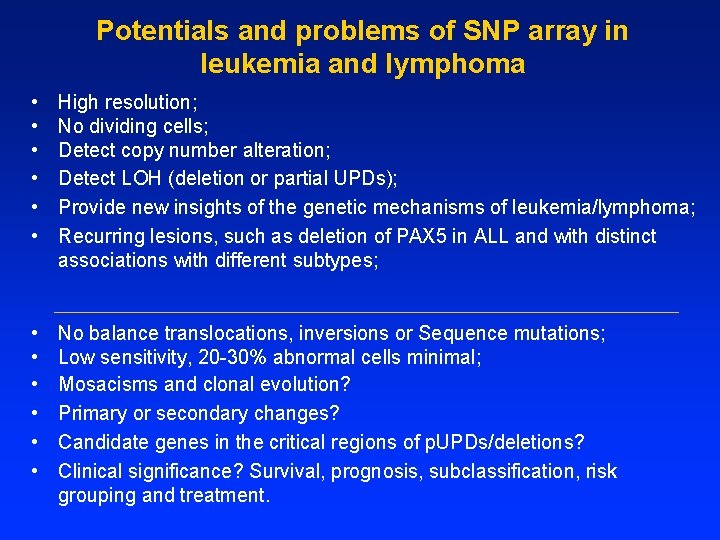
Potentials and problems of SNP array in leukemia and lymphoma • • • High resolution; No dividing cells; Detect copy number alteration; Detect LOH (deletion or partial UPDs); Provide new insights of the genetic mechanisms of leukemia/lymphoma; Recurring lesions, such as deletion of PAX 5 in ALL and with distinct associations with different subtypes; • • • No balance translocations, inversions or Sequence mutations; Low sensitivity, 20 -30% abnormal cells minimal; Mosacisms and clonal evolution? Primary or secondary changes? Candidate genes in the critical regions of p. UPDs/deletions? Clinical significance? Survival, prognosis, subclassification, risk grouping and treatment.

Can cytogenetics and FISH survive in the modern genomic era? Next-generation sequencing Thanks!


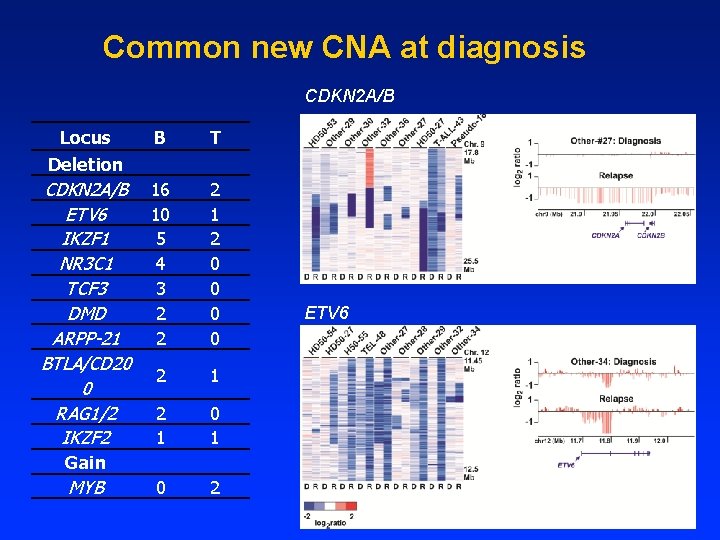
Common new CNA at diagnosis CDKN 2 A/B Locus Deletion B T CDKN 2 A/B ETV 6 IKZF 1 NR 3 C 1 TCF 3 DMD ARPP-21 BTLA/CD 20 0 RAG 1/2 IKZF 2 16 10 5 4 3 2 2 2 1 2 0 0 2 1 0 1 0 2 Gain MYB ETV 6

The power of SNP analysis in ALL • Genomic analyses provide new insights of the genetic mechanisms of ALL; • Recurring lesions, such as deletion of PAX 5, common in most subtypes of ALL and with distinct associations with different subtypes; • IKZF 1 alterations are a critical determinant of poor outcome; • Bioinformatics is critical to identify new therapeutic targets based on SNP data; • Existing analysis limited: copy number alteration, gene expression, limited sequencing;

Comparison of cytogenetics, FISH and SNP microarray Techniques Cytogenetics FISH SNP microarray Resolution + ++++ Sensitivity + +++ + neutral LOH – - + cell division + – – balanced lesions + - multiple clones + –/+ Screening for unknown defects + - + – +