Bulk RNAseq data Analysis From raw data to
Bulk RNA-seq data Analysis: From raw data to results to visualizations Skyler Kuhn 1, 2 Josh Meyer 1, 2 1. CCR Collaborative Bioinformatics Resource (CCBR), Center for Cancer Research, NCI 2. Advanced Biomedical Computational Science, Frederick National Laboratory for Cancer Research
1 Overview I. Bulk RNA-seq Pipeline Brief Introduction to Pipeliner Conceptual Overview of Workflow Importance of Quality-control and Quantification Pipeline

I. Bulk RNA-seq Pipeline
1 I. Brief Introduction to Pipeliner What is Pipeliner? ▫ TLDR: NGS pipelines developed and tested by experts at CCBR and NCBR What are the requirements? How do I access this platform? What skills do I need? ▫ Requires: Biowulf account ▫ Access: ssh -Y into Biowulf, grab an interactive node [optional], module load ccbrpipeliner ▫ User skillset: Basic understanding of unix-commands What is the starting point and end point(s) for Pipeliner’s RNA-seq pipeline? ▫ ▫ Starting Point: Fast. Q files (~raw data) Endpoint(s): Raw/normalized counts matrices, BAMs, Strand-specific Big. Wigs, Interactive Reports, and more… More information and documentation: ccbr. github. io/Pipeliner
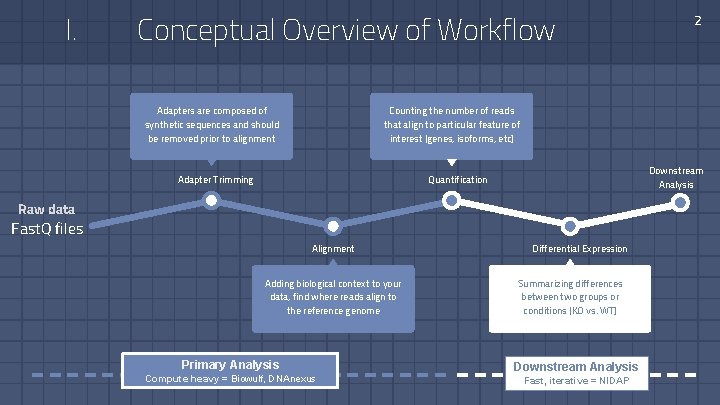
I. Conceptual Overview of Workflow Adapters are composed of synthetic sequences and should be removed prior to alignment 2 Counting the number of reads that align to particular feature of interest (genes, isoforms, etc) Downstream Analysis Quantification Adapter Trimming Raw data Fast. Q files Alignment Adding biological context to your data, find where reads align to the reference genome Primary Analysis Compute heavy = Biowulf, DNAnexus Differential Expression Summarizing differences between two groups or conditions (KO vs. WT) Downstream Analysis Fast, iterative = NIDAP
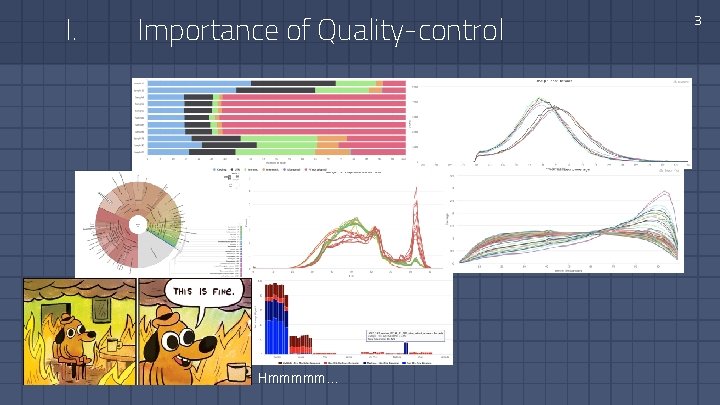
I. Importance of Quality-control Hmmmmm… 3

I. Quality-control and Quantification Pipeline Trim Align Count Cutadapt STAR 2 -pass RSEM Overview of Major Quality-control Steps ▫ Fast. QC ▫ Kraken and Fast. Q Screen Contamination Screening ▫ Samtools flagstat ▫ Quali. Map or BBtools Calculate insert size ▫ Preseq Estimate Library Complexity ▫ RSe. QC ▫ Picard ▫ Multi. QC Aggregates everything into an interactive report Assess sequencing quality Calculate alignment statistics Infer library stranded-ness, calculate TIN (~RIN) and read distribution over specific features, and much more… Calculate Duplication Rate, % reads in coding, intronic, intergenic regions, calculate Gene Coverage (5’/3’ bias) 4

I. Quality-control and Quantification Pipeline Trim Align Count Cutadapt STAR 2 -pass RSEM Overview of Major Quality-control Steps ▫ Fast. QC ▫ Kraken and Fast. Q Screen Contamination Screening ▫ Samtools flagstat ▫ Quali. Map or BBtools Calculate insert size ▫ Preseq Estimate Library Complexity ▫ RSe. QC ▫ Picard ▫ Multi. QC Aggregates everything into an interactive report Assess sequencing quality Calculate alignment statistics Infer library stranded-ness, calculate TIN (~RIN) and read distribution over specific features, and much more… Calculate Duplication Rate, % reads in coding, intronic, intergenic regions, calculate Gene Coverage (5’/3’ bias) 4
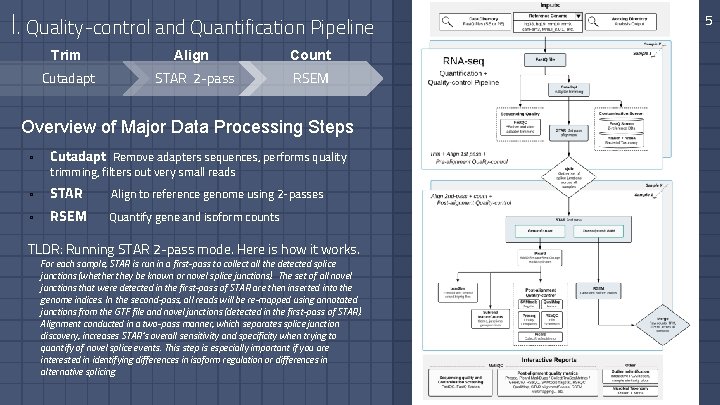
I. Quality-control and Quantification Pipeline Trim Align Count Cutadapt STAR 2 -pass RSEM Overview of Major Data Processing Steps ▫ Cutadapt Remove adapters sequences, performs quality ▫ STAR Align to reference genome using 2 -passes ▫ RSEM Quantify gene and isoform counts trimming, filters out very small reads TLDR: Running STAR 2 -pass mode. Here is how it works. For each sample, STAR is run in a first-pass to collect all the detected splice junctions (whether they be known or novel splice junctions). The set of all novel junctions that were detected in the first-pass of STAR are then inserted into the genome indices. In the second-pass, all reads will be re-mapped using annotated junctions from the GTF file and novel junctions (detected in the first-pass of STAR). Alignment conducted in a two-pass manner, which separates splice junction discovery, increases STAR’s overall sensitivity and specificity when trying to quantify of novel splice events. This step is especially important if you are interested in identifying differences in isoform regulation or differences in alternative splicing. 5
THANKS! Acknowledgements CCBR, NCBR, and GAU members Any questions?
- Slides: 10