Broad Course Objectives Review the structure of amino

Broad Course Objectives • Review the structure of amino acids and polypeptide chains; • Understand how gene sequence determines amino acid sequence • Explain the structure and function of operons; • Explain the structure and function of ribosomes Necessary for material on Mutations and Hemoglobin Analysis lab.

Study Guide/Outline--Translation Basic structure of protein • What is the amino-end and the carboxy-end of a polypeptide chain (amino acid chain)? How do the amino acids differ from one another? • What is a peptide bond? What is the difference between 1 o, 2 o, 3 o and 4 o structure in proteins? Deciphering the m. RNA Transcript • Be able to predict RNA transcript and amino-acid chains if given the sequence of DNA and the codon table. • How does the sequence of DNA nucleotides specify the sequence of amino acids in the protein for which it codes? • What is a codon? What is an anti-codon and where is it found? What are “Start” and “Stop” codons? • Does every codon correspond to different amino acids? Which nucleotide within the codon [1 st, 2 nd, or 3 rd] varies the most and still specifies the same amino acid? • In general, what roles do t. RNAs and ribosomes play in translation? How are t. RNA’s “charged”? How is m. RNA Translated? • What are the events associated with Initiation, Elongation, and Termination during translation? • What does each component of the translation complex—m. RNA, t. RNA, small ribosomal subunit, large ribosomal subunit—do during translation? • Which subunit joins the amino acids together? Why is it called a “ribozyme”? • What is a “reading frame”? • How does translation end? • In what part of the cell does translation occur?
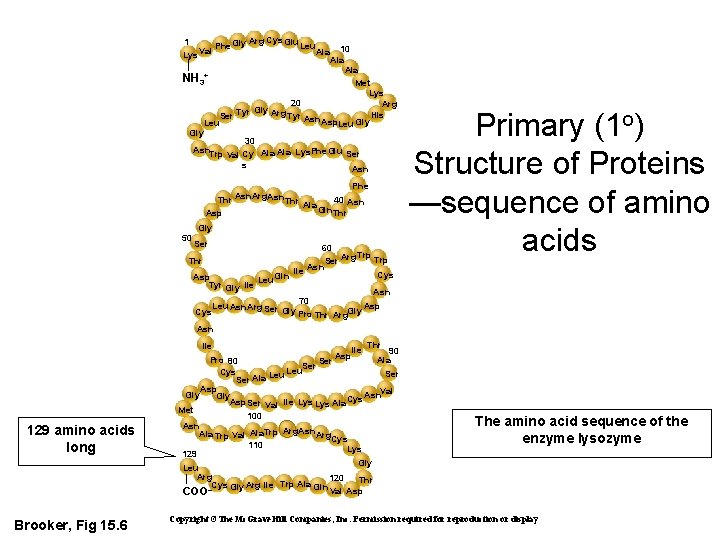
Arg Cys Glu 1 Phe Gly Leu Val Ala Lys 10 Ala NH 3+ Met Lys 20 Arg Gly Arg His Tyr Asn Ser Tyr Asp. Leu Gly 30 Asn. Trp Val Cy Ala Lys. Phe Glu Ser s Asn Thr Asn Asp Arg. Asn Thr Phe 40 Asn Ala Gin Thr Gly 50 Ser 60 Trp Ser Arg Asn lle Cys Gln Asp Tyr Gly lle Leu Asn 70 Asp Leu Asn Arg Ser Gly Pro Thr Arg. Gly Cys Thr Primary (1 o) Structure of Proteins —sequence of amino acids Asn lle Thr 90 Asp Ala Pro 80 Ser Leu Cys Ser Leu Ser Ala Asp Gly Asn. Val Gly Asp. Ser Val lle Lys Ala Cys Met 100 Asn Ala Trp Val Ala. Trp Arg. Asn Arg. Cys 110 Lys 129 Gly Leu Arg 120 Thr Cys Gly Arg lle Trp Ala Gln Val Asp COO– lle 129 amino acids long Brooker, Fig 15. 6 The amino acid sequence of the enzyme lysozyme Copyright ©The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display
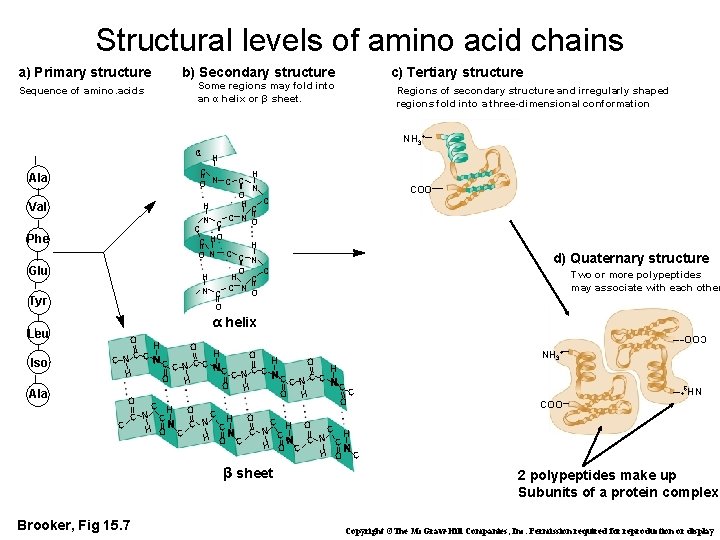
Structural levels of amino acid chains a) Primary structure c) Tertiary structure b) Secondary structure Some regions may fold into an α helix or β sheet. Sequence of amino. acids Regions of secondary structure and irregularly shaped regions fold into a three-dimensional conformation NH 3+ C Ala Val Phe Glu Tyr Ala d) Quaternary structure Two or more polypeptides may associate with each other O α helix O CNCC H H O CCNCC O H H O CCN CC O H H O O CC N C C O H NH 3+ H C O C C H O C C N C C H O N CH O C C H C N C H O C β sheet Brooker, Fig 15. 7 NH 3+ Iso COO– Leu H C H N C C O N O C H H C N C C NO C H H O N C C N C O H H C N C C NO COO– 2 polypeptides make up Subunits of a protein complex Copyright ©The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display
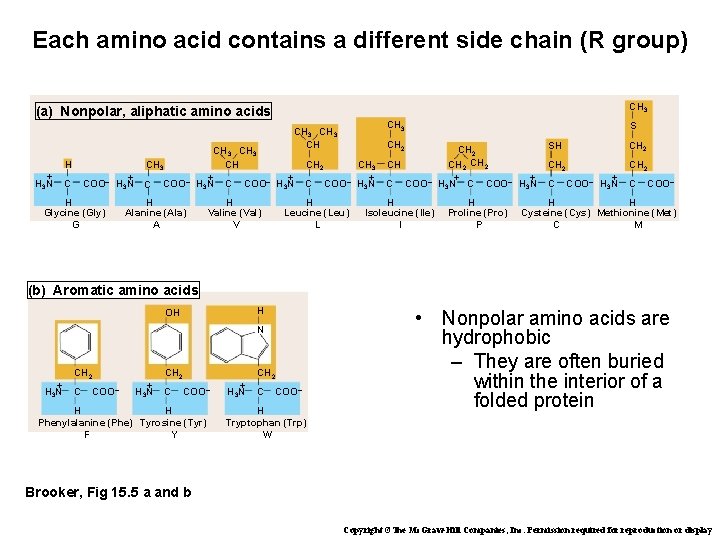
Each amino acid contains a different side chain (R group) CH 3 (a) Nonpolar, aliphatic amino acids CH 3 CH H + H 3 N COO– C + H 3 N H Glycine (Gly) G CH 3 CH + + C COO– H 3 N H Alanine (Ala) A H Valine (Val) V CH 2 C S COO– H Leucine (Leu) L CH 3 + H 3 N CH C COO– H Isoleucine (Ile) I CH 2 + + H 3 N C COO– H 3 N H Proline (Pro) P CH 2 SH CH 2 C COO– + H 3 N CH 2 C COO– H H Cysteine (Cys) Methionine (Met) C M (b) Aromatic amino acids H OH N + H 3 N CH 2 C COO– H H Phenylalanine (Phe) Tyrosine (Tyr) F Y + H 3 N CH 2 C COO– H Tryptophan (Trp) W • Nonpolar amino acids are hydrophobic – They are often buried within the interior of a folded protein Brooker, Fig 15. 5 a and b Copyright ©The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display
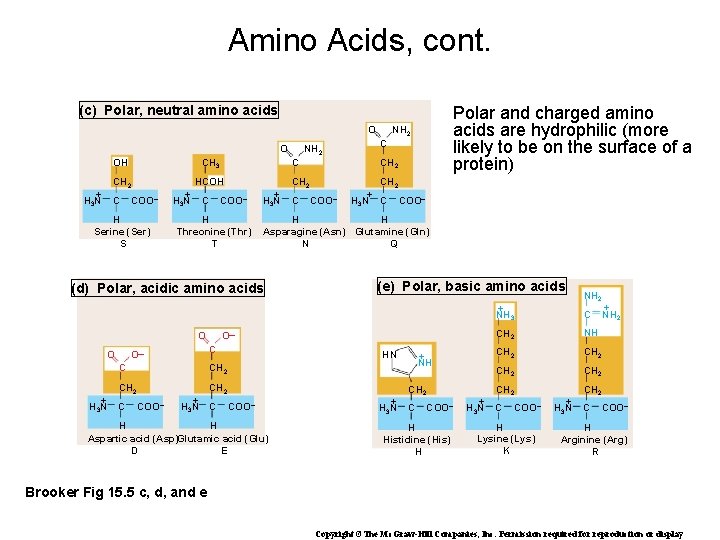
Amino Acids, cont. (c) Polar, neutral amino acids O O HCOH CH 2 + H 3 N C COO– H Serine (Ser) S + H 3 N C COO– H Threonine (Thr) T + H 3 N C C CH 2 C COO– + H 3 N COO– CH 2 + H 3 N C COO– (e) Polar, basic amino acids O– O O– CH 2 C NH 2 C H H Asparagine (Asn) Glutamine (Gln) N Q (d) Polar, acidic amino acids O NH 2 C CH 3 OH Polar and charged amino acids are hydrophilic (more likely to be on the surface of a protein) C HN COO– H H Aspartic acid (Asp)Glutamic acid (Glu) D E + H 3 N + NH CH 2 C COO– H Histidine (His) H + H 3 N NH 2 + NH 3 C CH 2 NH CH 2 CH 2 C COO– H Lysine (Lys) K + H 3 N + NH 2 C COO– H Arginine (Arg) R Brooker Fig 15. 5 c, d, and e Copyright ©The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display
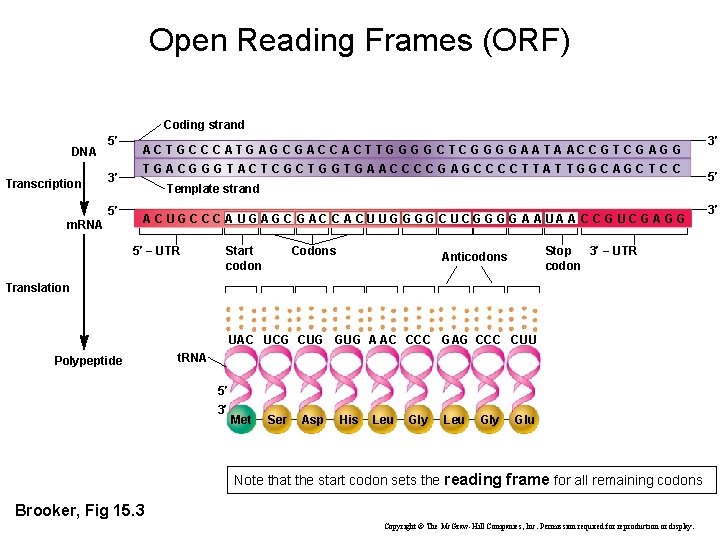
Open Reading Frames (ORF) Coding strand DNA Transcription 5′ 3′ 5′ m. RNA A C T G C C C A T G A G C G A C C A C T T G G C T C G G A A T A AC C G T C G A G G T G AC G G G T AC T C G C T G G T G A AC C G AG C C T T AT T G GC AG C T C C Template strand A C U G C C C A U G A G C G AC C A C U U G G C U C G G A A U A A C C G U C G A G G 5′ − UTR Start codon Codons Stop 3′ − UTR codon Anticodons Translation UAC UCG CUG GUG A AC CCC GAG CCC CUU Polypeptide t. RNA 5′ 3′ Met Ser Asp His Leu Gly Glu Note that the start codon sets the reading frame for all remaining codons Brooker, Fig 15. 3 Copyright © The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display. 3′ 5′ 3′
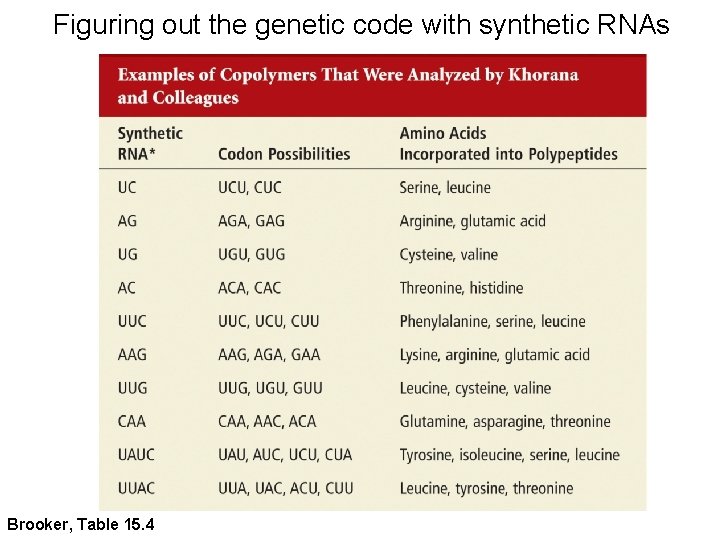
Figuring out the genetic code with synthetic RNAs Brooker, Table 15. 4
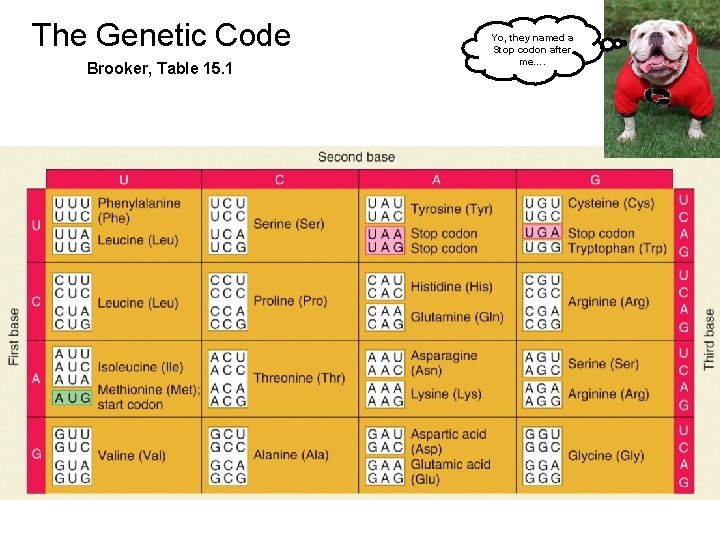
The Genetic Code Brooker, Table 15. 1 Yo, they named a Stop codon after me….

Published gene sequences are the nontemplate strand for ease of translating the sequence into protein • http: //www. ncbi. nlm. nih. gov/entrez/quer y. fcgi? db=OMIM
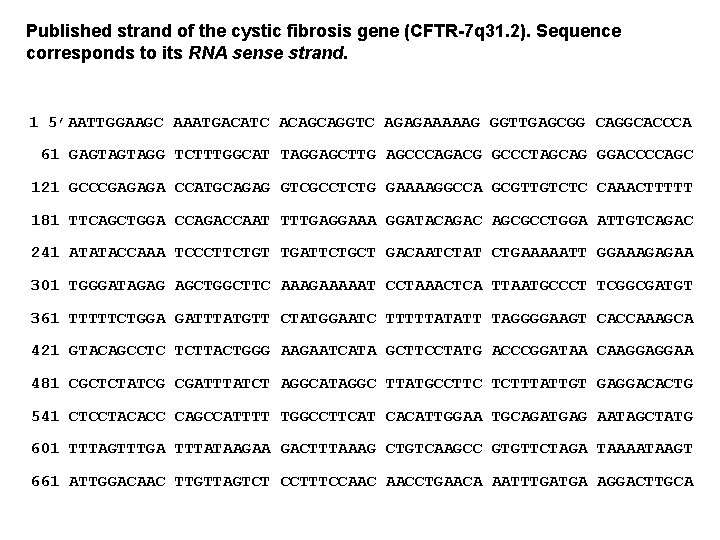
Published strand of the cystic fibrosis gene (CFTR-7 q 31. 2). Sequence corresponds to its RNA sense strand. 1 5’AATTGGAAGC AAATGACATC ACAGCAGGTC AGAGAAAAAG GGTTGAGCGG CAGGCACCCA 61 GAGTAGTAGG TCTTTGGCAT TAGGAGCTTG AGCCCAGACG GCCCTAGCAG GGACCCCAGC 121 GCCCGAGAGA CCATGCAGAG GTCGCCTCTG GAAAAGGCCA GCGTTGTCTC CAAACTTTTT 181 TTCAGCTGGA CCAGACCAAT TTTGAGGAAA GGATACAGAC AGCGCCTGGA ATTGTCAGAC 241 ATATACCAAA TCCCTTCTGT TGATTCTGCT GACAATCTAT CTGAAAAATT GGAAAGAGAA 301 TGGGATAGAG AGCTGGCTTC AAAGAAAAAT CCTAAACTCA TTAATGCCCT TCGGCGATGT 361 TTTTTCTGGA GATTTATGTT CTATGGAATC TTTTTATATT TAGGGGAAGT CACCAAAGCA 421 GTACAGCCTC TCTTACTGGG AAGAATCATA GCTTCCTATG ACCCGGATAA CAAGGAGGAA 481 CGCTCTATCG CGATTTATCT AGGCATAGGC TTATGCCTTC TCTTTATTGT GAGGACACTG 541 CTCCTACACC CAGCCATTTT TGGCCTTCAT CACATTGGAA TGCAGATGAG AATAGCTATG 601 TTTAGTTTGA TTTATAAGAA GACTTTAAAG CTGTCAAGCC GTGTTCTAGA TAAAATAAGT 661 ATTGGACAAC TTGTTAGTCT CCTTTCCAAC AACCTGAACA AATTTGATGA AGGACTTGCA
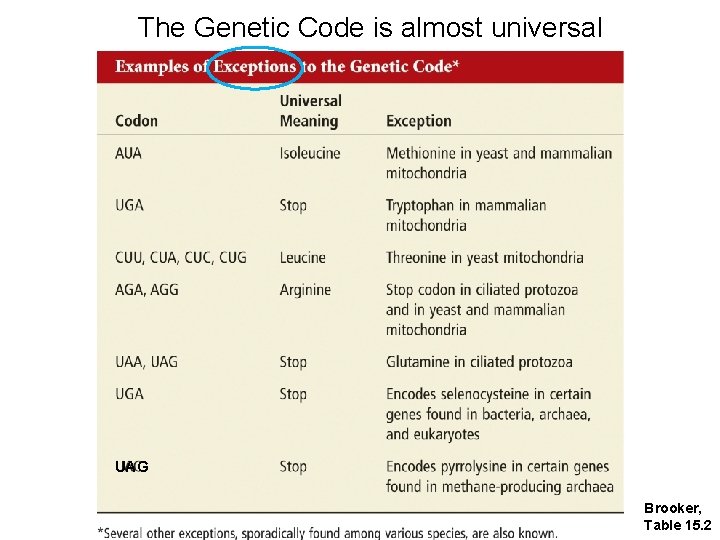
The Genetic Code is almost universal UAG Brooker, Table 15. 2
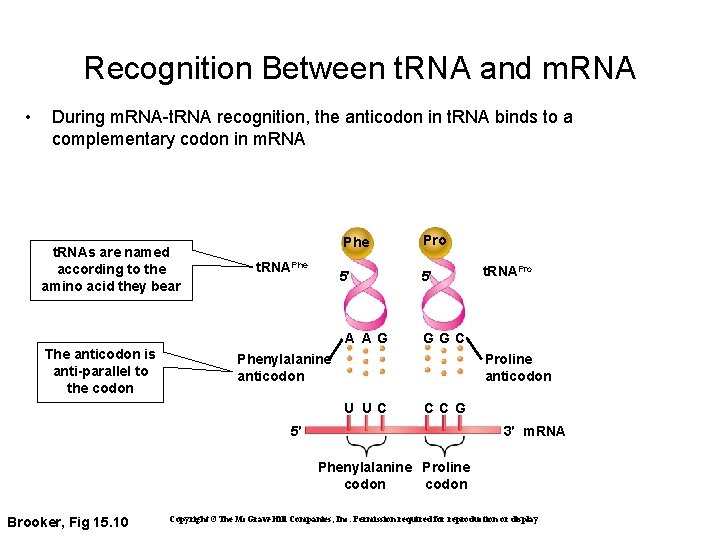
Recognition Between t. RNA and m. RNA • During m. RNA-t. RNA recognition, the anticodon in t. RNA binds to a complementary codon in m. RNA t. RNAs are named according to the amino acid they bear t. RNAPhe Pro 5′ 5′ A AG The anticodon is anti-parallel to the codon t. RNAPro GGC Phenylalanine anticodon Proline anticodon U UC CCG 5′ 3′ m. RNA Phenylalanine Proline codon Brooker, Fig 15. 10 Copyright ©The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display
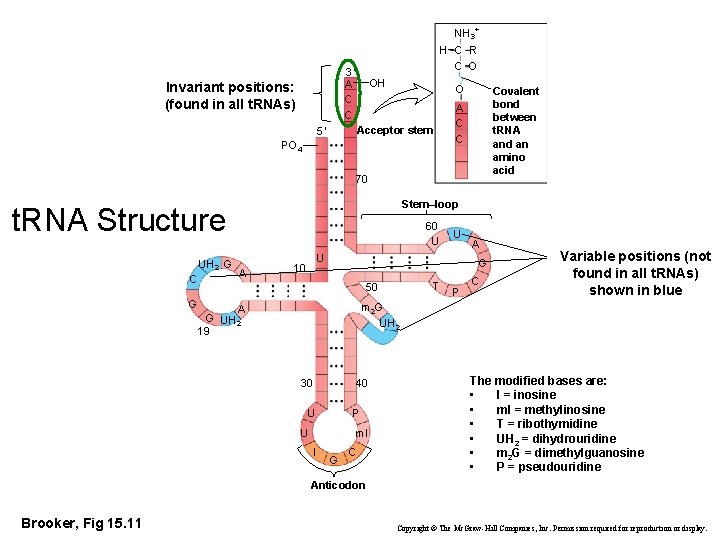
NH 3+ H C R C O 3′ A C C Invariant positions: (found in all t. RNAs) OH O Acceptor stem 5′ PO 4 Covalent bond between t. RNA and an amino acid A C C 70 Stem–loop t. RNA Structure UH 2 G C 60 U A U U 10 A G T 50 G P C Variable positions (not found in all t. RNAs) shown in blue m 2 G UH 2 A G UH 2 19 40 30 U P U m. I I G C The modified bases are: • I = inosine • m. I = methylinosine • T = ribothymidine • UH 2 = dihydrouridine • m 2 G = dimethylguanosine • P = pseudouridine Anticodon Brooker, Fig 15. 11 Copyright © The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display.
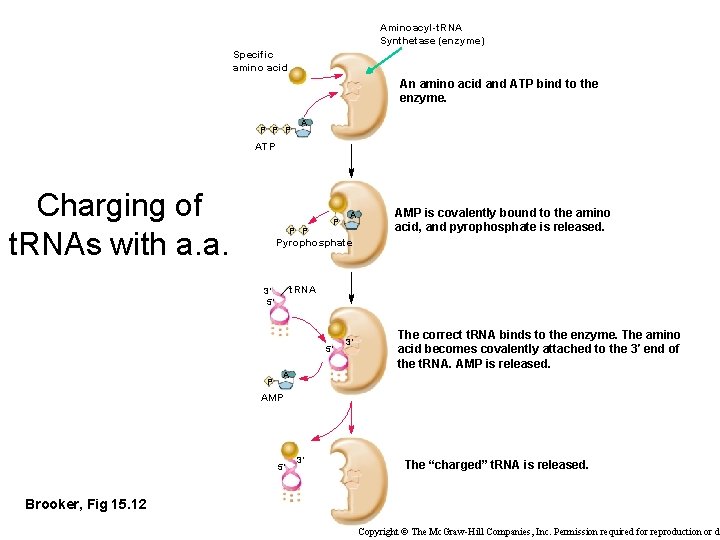
Aminoacyl-t. RNA Synthetase (enzyme) Specific amino acid An amino acid and ATP bind to the enzyme. P P P A ATP Charging of t. RNAs with a. a. P P P A AMP is covalently bound to the amino acid, and pyrophosphate is released. Pyrophosphate t. RNA 3′ 5′ 5′ P A 3′ The correct t. RNA binds to the enzyme. The amino acid becomes covalently attached to the 3′ end of the t. RNA. AMP is released. AMP 5′ 3′ The “charged” t. RNA is released. Brooker, Fig 15. 12 Copyright © The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or d
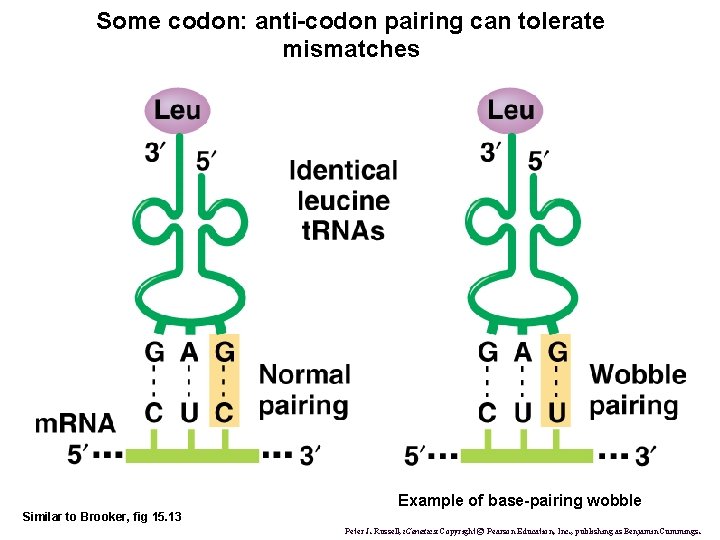
Some codon: anti-codon pairing can tolerate mismatches Example of base-pairing wobble Similar to Brooker, fig 15. 13 Peter J. Russell, i. Genetics: Copyright © Pearson Education, Inc. , publishing as Benjamin Cummings.
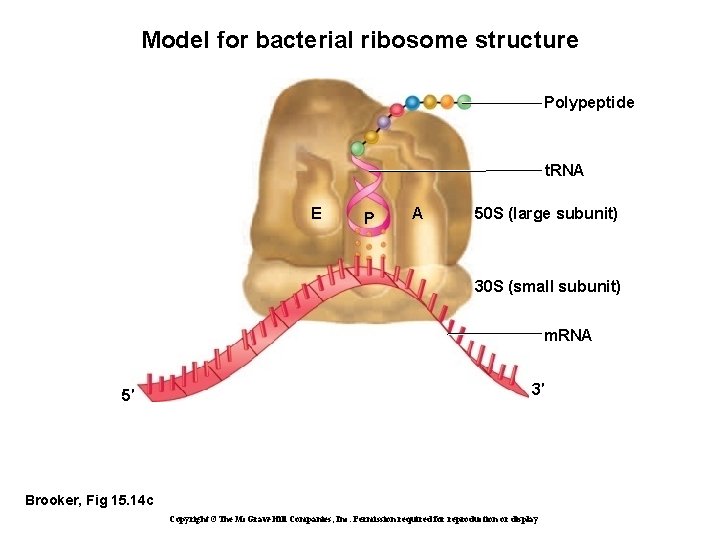
Model for bacterial ribosome structure Polypeptide t. RNA E P A 50 S (large subunit) 30 S (small subunit) m. RNA 5′ 3′ Brooker, Fig 15. 14 c Copyright ©The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display
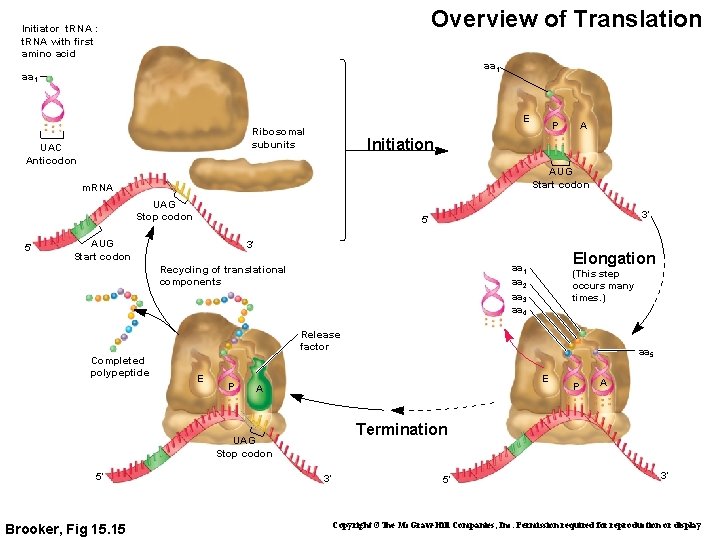
Overview of Translation Initiator t. RNA : t. RNA with first amino acid aa 1 E Ribosomal subunits UAC Anticodon A Initiation AUG Start codon m. RNA UAG Stop codon 5′ P 3′ 5′ AUG Start codon 3′ Elongation aa 1 aa 2 aa 3 aa 4 Recycling of translational components (This step occurs many times. ) Release factor Completed polypeptide E P E A Brooker, Fig 15. 15 P A Termination UAG Stop codon 5′ aa 5 3′ 5′ 3′ Copyright ©The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display
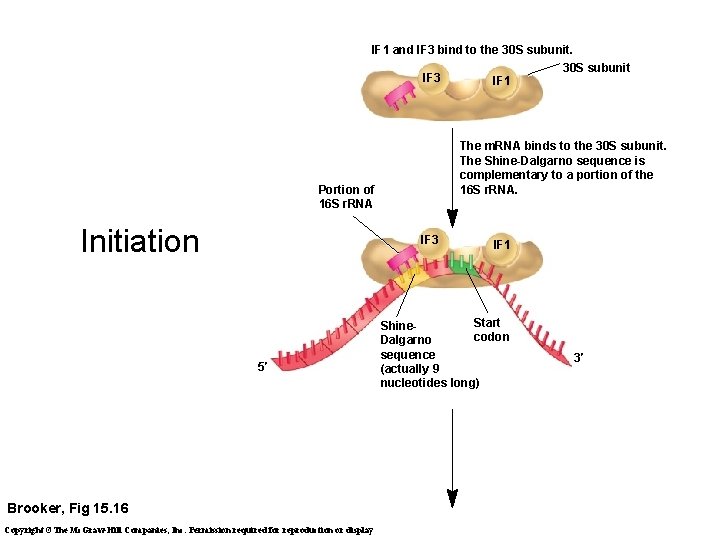
IF 1 and IF 3 bind to the 30 S subunit. IF 3 The m. RNA binds to the 30 S subunit. The Shine-Dalgarno sequence is complementary to a portion of the 16 S r. RNA. Portion of 16 S r. RNA Initiation IF 3 5′ Brooker, Fig 15. 16 Copyright ©The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display IF 1 30 S subunit IF 1 Start Shinecodon Dalgarno sequence (actually 9 nucleotides long) 3′
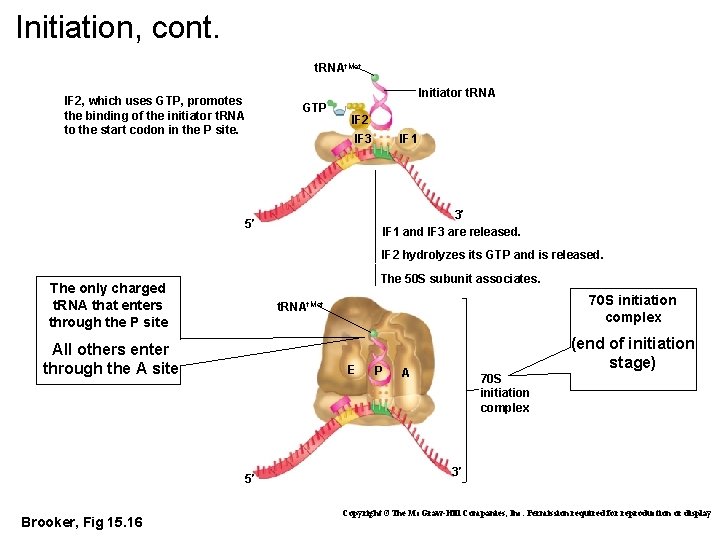
Initiation, cont. t. RNAf. Met IF 2, which uses GTP, promotes the binding of the initiator t. RNA to the start codon in the P site. Initiator t. RNA GTP IF 2 IF 1 IF 3 3′ IF 1 and IF 3 are released. 5′ IF 2 hydrolyzes its GTP and is released. The 50 S subunit associates. The only charged t. RNA that enters through the P site All others enter through the A site E 5′ Brooker, Fig 15. 16 70 S initiation complex t. RNAf. Met P (end of initiation stage) A 70 S initiation complex 3′ Copyright ©The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display
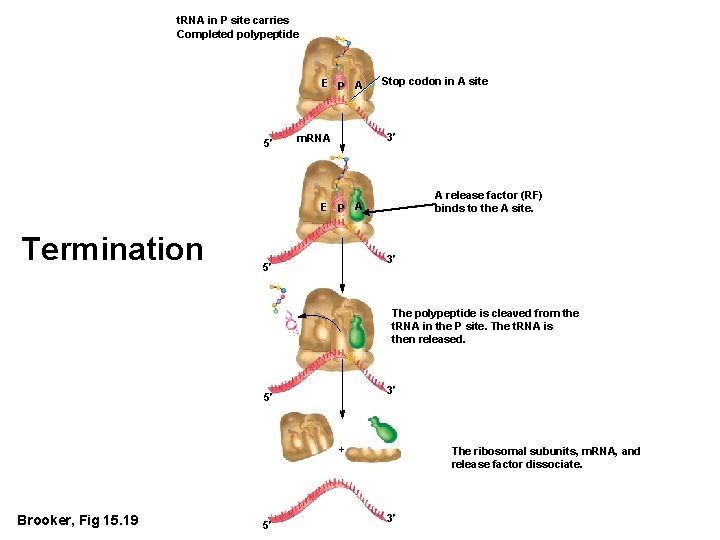
t. RNA in P site carries Completed polypeptide E P A 5′ Stop codon in A site 3′ m. RNA A release factor (RF) binds to the A site. E P A Termination 3′ 5′ The polypeptide is cleaved from the t. RNA in the P site. The t. RNA is then released. 3′ 5′ + Brooker, Fig 15. 19 5′ The ribosomal subunits, m. RNA, and release factor dissociate. 3′

Animation of translation http: //vcell. ndsu. nodak. edu/animations/

Predict the amino acid sequence produced during translation by the following short theoretical m. RNA sequences. Note that the second sequence was formed from the first by a deletion of only one nucleotide. • AUG CCG GAU UAU AGU UGA • AUG CCG GAU UAA GUU GA

Predict the amino acid sequence produced during translation by the following short theoretical m. RNA sequences. Note that the second sequence was formed from the first by a deletion of only one nucleotide. • AUG CCG GAU UAU AGU UGA • Met Pro Asp Tyr Ser STOP • AUG CCG GAU UAA GUU GA • Met Pro Asp STOP
- Slides: 24