Biopackages net Operating System Packages for Bioinformatics Allen

Biopackages. net Operating System Packages for Bioinformatics Allen Day 2005. 17
What is a package? o o Software, config files, documentation, and/or data encapsulated in a single file Metadata describing: n n n Version, license, package “category” Dependencies What the package provides

o GMOD target audience n Small MODs
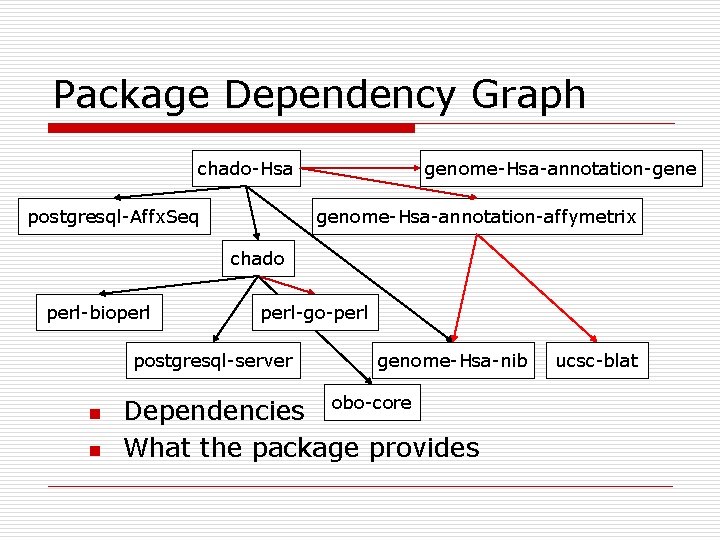
Package Dependency Graph chado-Hsa postgresql-Affx. Seq genome-Hsa-annotation-gene genome-Hsa-annotation-affymetrix chado perl-bioperl-go-perl postgresql-server n n genome-Hsa-nib Dependencies obo-core What the package provides ucsc-blat
Dependencies o o Build Dependency Installation Dependency

What is a Package Manager? o Tools to manage installation, upgrade, uninstallation of packages n n Verify package integrity (checksums) Maintain system integrity o o n n n Transactional Allow rollbacks Dependency checking Dependency graph recursion Allow software customization (patches)
Why bioinformatics packages? o Consistency of installation process n o Automatic dependency installation n o Perl modules especially bad – bioperl has 60+ modules in its dependency tree Integrity/Auditing of system state n o Bioinfo. package installs vary wildly, and commonly lack documentation Know an installed package works, which version, how to replicate system setup Tighter integration with operating system n Daemons, config & log file locations, etc.

What’s available? o RPM packages only right now n o Primary focus on Fedora Core 2 Some RPMs also available for n n n Fedora Core 3 Red. Hat 9 Cygwin
What’s available? o Three primary foci n n n Applications Libraries Data sets

Applications o o o Gbrowse Textpresso BLAT daemon NCBI Toolkit (BLAST, etc) HMMer
What’s available? o Libraries n n Bioperl R & Bioconductor Squid EMBOSS
What’s available? o Data sets n n Genome & protein sequence Sequence features Ontologies All installed using a common directory structure
What’s available? o o o UCSC tools (utilities, BLAT system service, CGI scripts) Bioperl R / Bioconductor GMOD apps (Gbrowse, Textpresso, …) Data packages n n Genome sequence (fa, nib, blastdb) Genome features (Affy probeset alignments, m. RNA, etc)
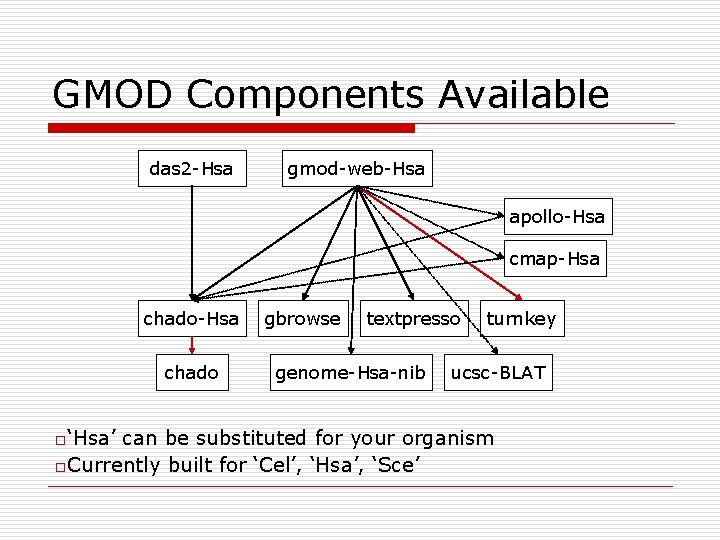
GMOD Components Available das 2 -Hsa gmod-web-Hsa apollo-Hsa cmap-Hsa chado o‘Hsa’ gbrowse textpresso genome-Hsa-nib turnkey ucsc-BLAT can be substituted for your organism o. Currently built for ‘Cel’, ‘Hsa’, ‘Sce’
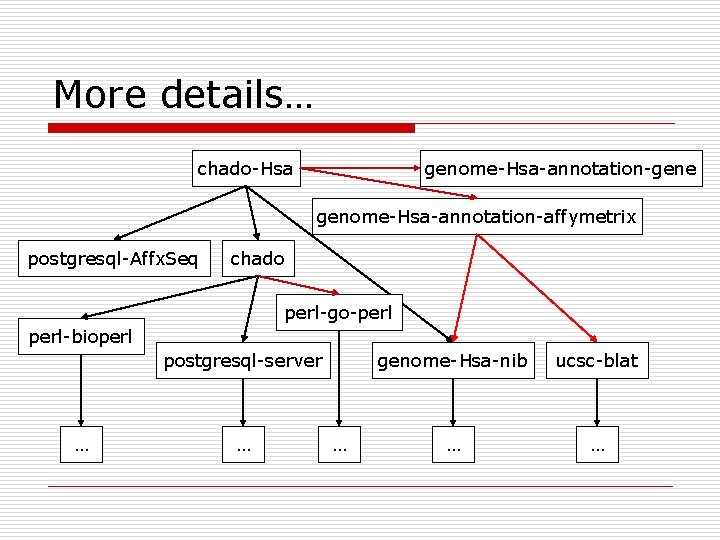
More details… chado-Hsa genome-Hsa-annotation-gene genome-Hsa-annotation-affymetrix postgresql-Affx. Seq chado perl-go-perl-bioperl postgresql-server … … … genome-Hsa-nib ucsc-blat … …
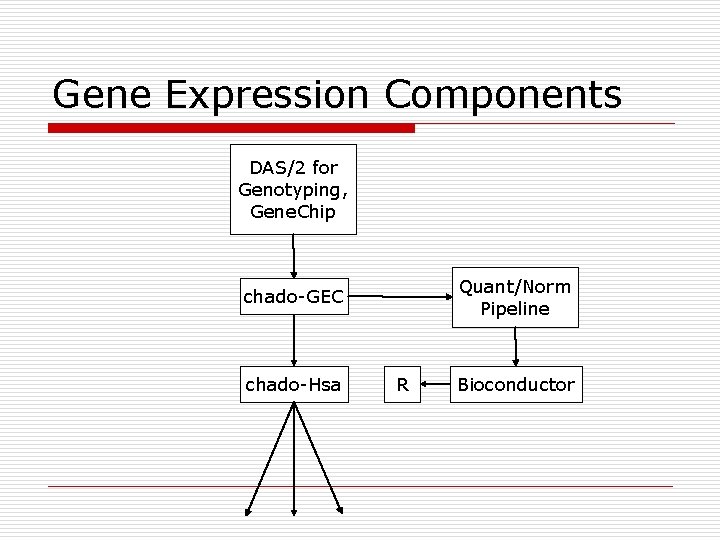
Gene Expression Components DAS/2 for Genotyping, Gene. Chip Quant/Norm Pipeline chado-GEC chado-Hsa R Bioconductor

Resources o http: //www. biopackages. net n n ~1000 RPMs for Fedora Core 2, 3 Available via yum o See site for a configuration example.
TODO o Support more architectures n o Automate package build process n o Build for Cygwin & OS X. RPM has been ported to both Build farm of multiple architectures, controllable via scheduler (Grid. Engine) Automate (if possible) inclusion of new software / data releases

TODO o Build community interest and involvement n n Keep adding more packages! Keep existing packages current!
Acknowledgements o o o Patrick Alger Jared Fox Brian O’Connor Todd Harris Lincoln Stein Stanley Nelson
- Slides: 20