Biomolecular NMR Spectroscopy Methods and applications to proteins

Biomolecular NMR Spectroscopy Methods and applications to proteins Robert Kaptein Novosibirsk, November 2012 1

Contents - Introduction to Biomolecular NMR Multidimensional NMR for proteins Resonance assignments Observables & structure restraints 3 D Structure determination by NMR Protein Dynamics Example: Lac repressor, structure, dynamics, DNA interactions 2

Introduction to Biomolecular NMR 3

Elements of Protein NMR 4
5 Pros & cons of NMR in structural biology • Pros. . . – no need for crystal: • no crystal packing artifacts, solution more nativelike – study of dynamics: • picosecond to seconds time scales, conformational averaging, chemical reactions, folding. . . – easy study of protein-protein, protein-DNA, proteinligand interactions • Cons. . . – NMR structure determination is a bit slow. . – Need isotope labeling (13 C, 15 N) – solution NMR works best for MW < 50 k. Da
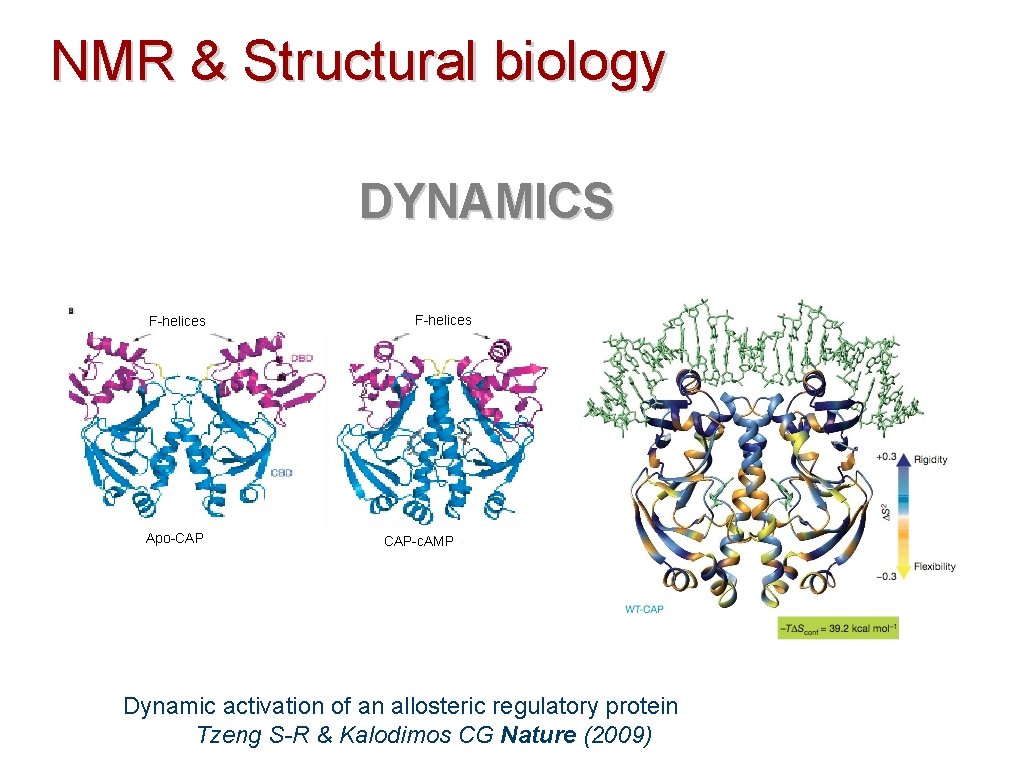
NMR & Structural biology DYNAMICS F-helices Apo-CAP F-helices CAP-c. AMP Dynamic activation of an allosteric regulatory protein Tzeng S-R & Kalodimos CG Nature (2009)

NMR & Structural biology TRANSIENT COMPLEXES Visualization of the Encounter Ensemble of the Transient Electron Transfer Complex of Cytochrome c and Cytochrome c Peroxidase Bashir Q. et al JACS (2010) 7
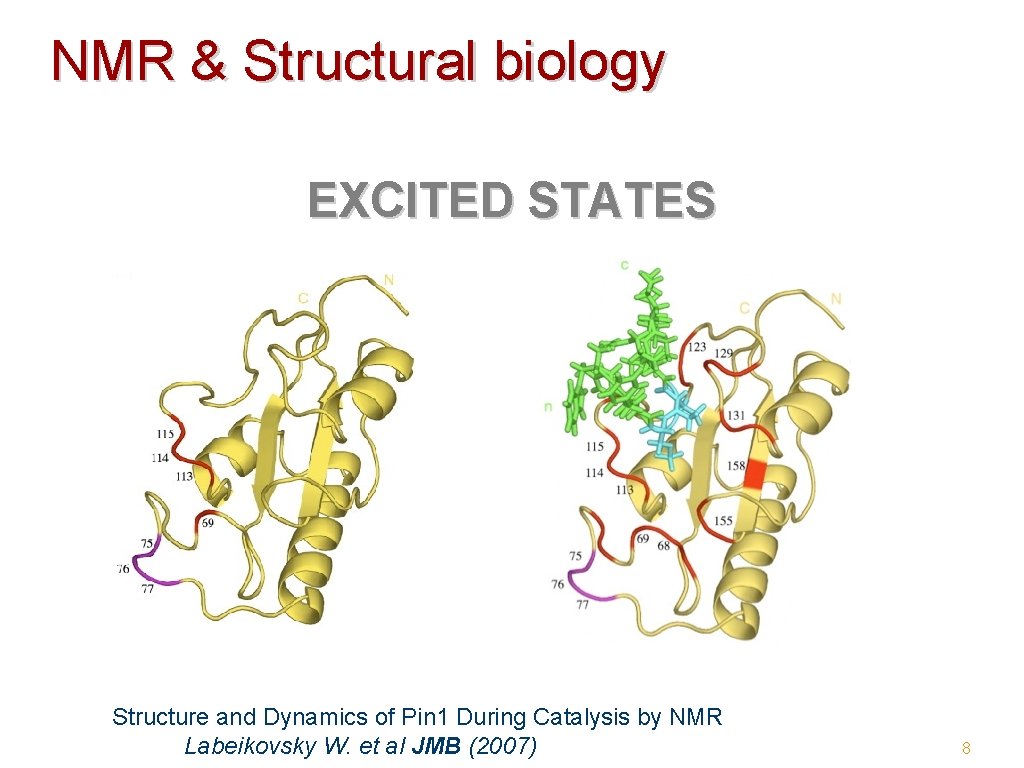
NMR & Structural biology EXCITED STATES Structure and Dynamics of Pin 1 During Catalysis by NMR Labeikovsky W. et al JMB (2007) 8
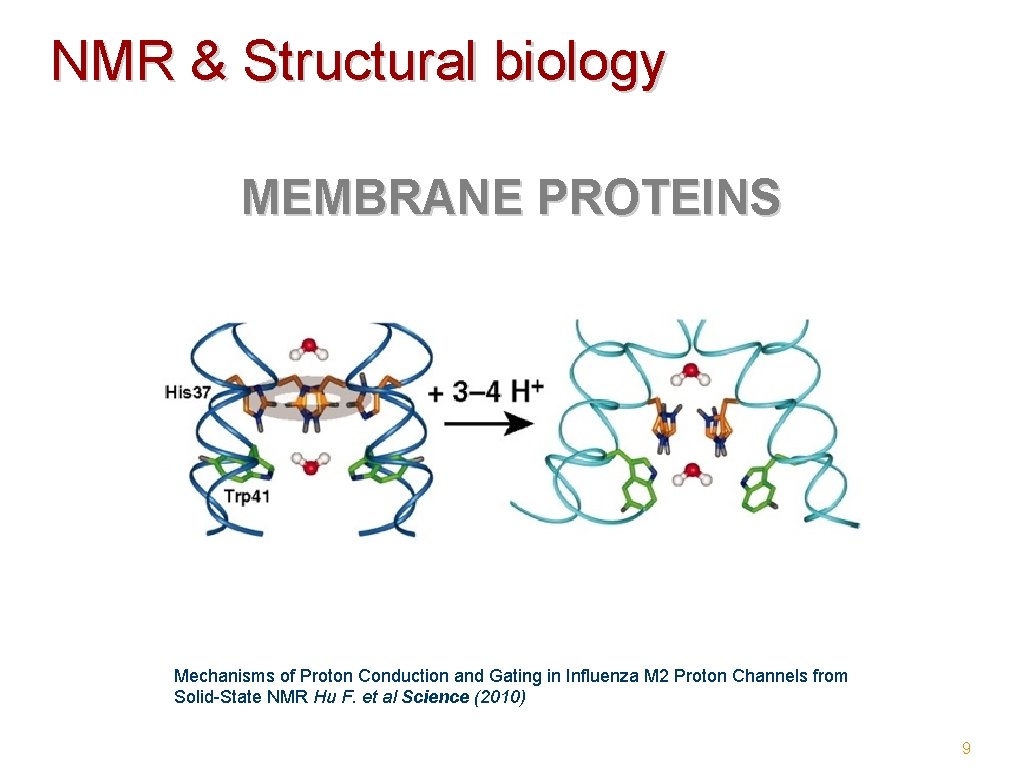
NMR & Structural biology MEMBRANE PROTEINS Mechanisms of Proton Conduction and Gating in Influenza M 2 Proton Channels from Solid-State NMR Hu F. et al Science (2010) 9
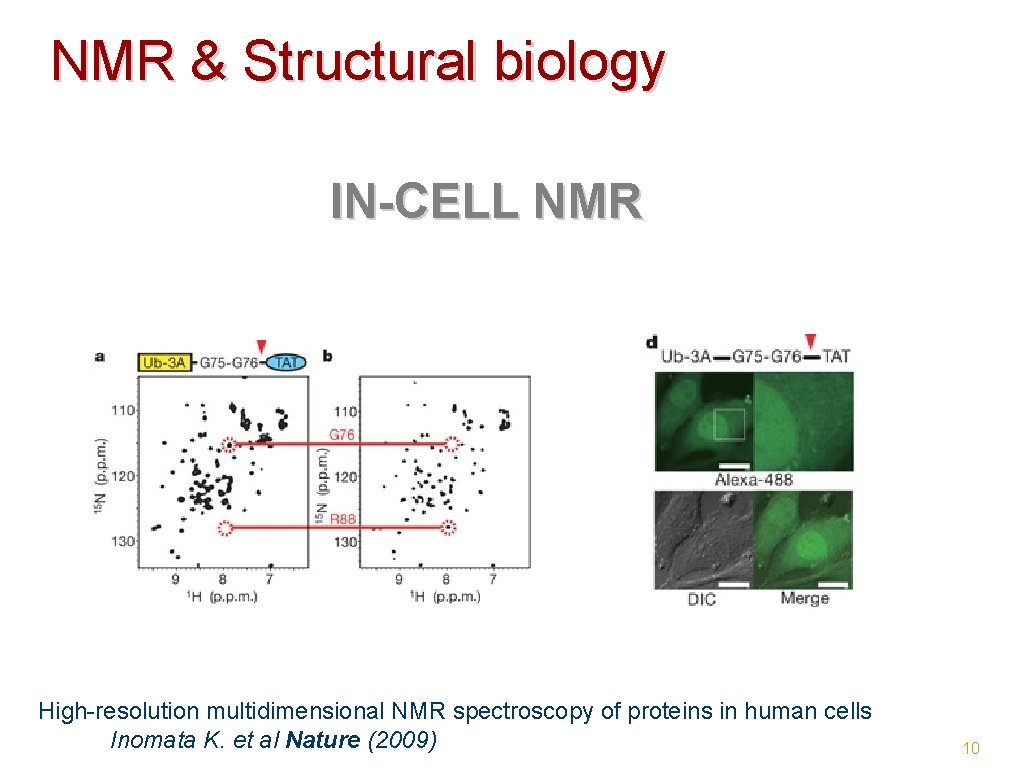
NMR & Structural biology IN-CELL NMR High-resolution multidimensional NMR spectroscopy of proteins in human cells Inomata K. et al Nature (2009) 10
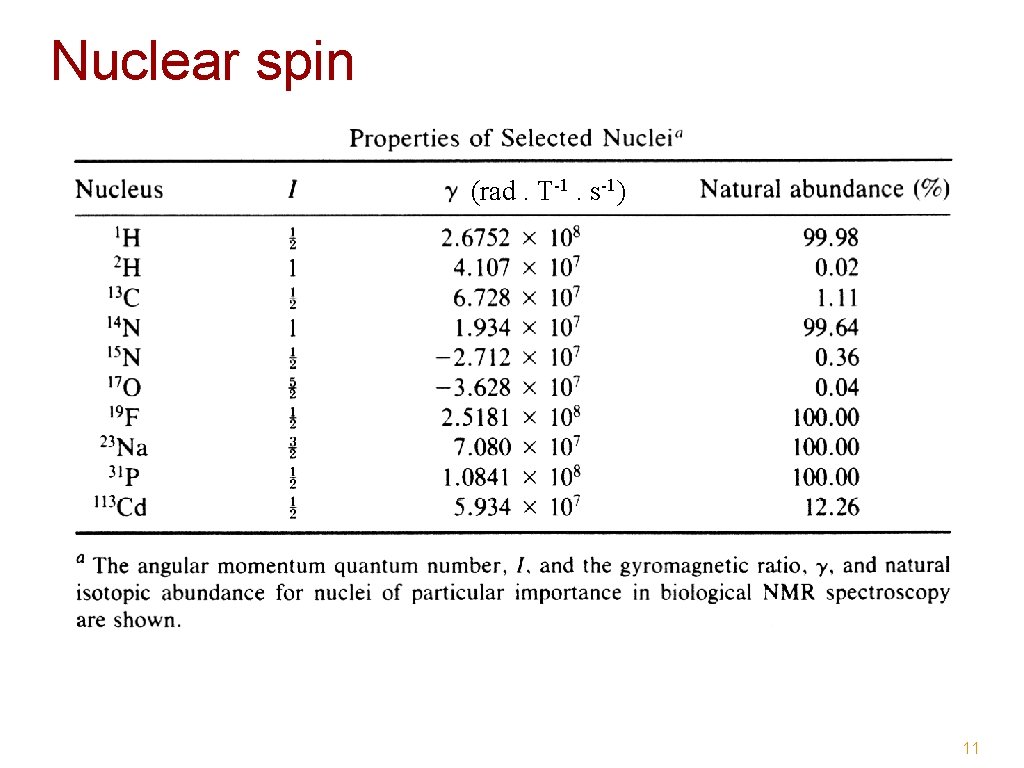
Nuclear spin (rad. T-1. s-1) 11
The NMR sample • Isotope labeling - - 15 N for small proteins < 15 k. Da 15 N & 13 C for larger proteins, up to 30 -40 k. Da 15 N, 13 C & 2 H for large proteins > 40 k. Da selective labeling (e. g. only methyl groups) • Sample - recombinant expression in E. coli - pure, stable and high concentration ➡ 500 µL of 0. 5 m. M solution: ~ 5 mg per sample preferably low salt, low p. H 12
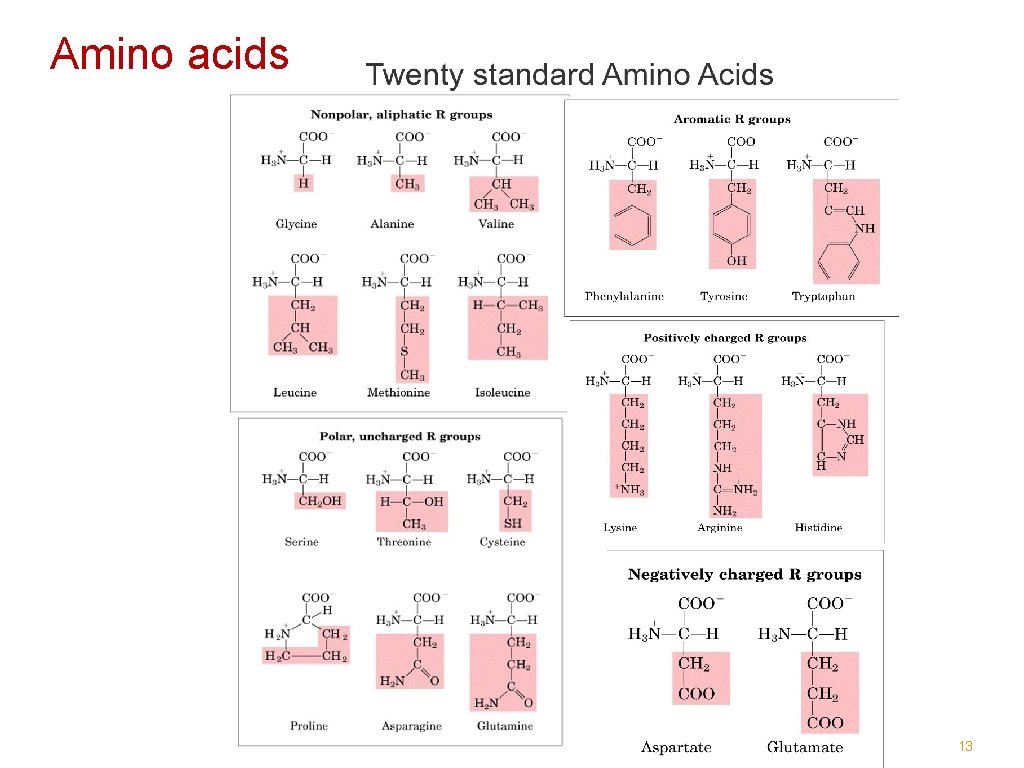
Amino acids 13

Amino acids are usually referred to with either a three-letter or a one-letter code: Glycine Gly G Histidine His H Alanine Ala A Proline Pro P Valine Val V Aspartate Asp D Leucine Leu L Glutamate Glu E Isoleucine Ile I Asparagine Asn N Serine Ser S Glutamine Gln Q Threonine Thr T Lysine Lys K Phenylalanine Phe F Arginine Arg R Tyrosine Tyr Y Cysteine Cys C Tryptophane Trp W Methionine Met M

Multidimensional NMR for Proteins 15
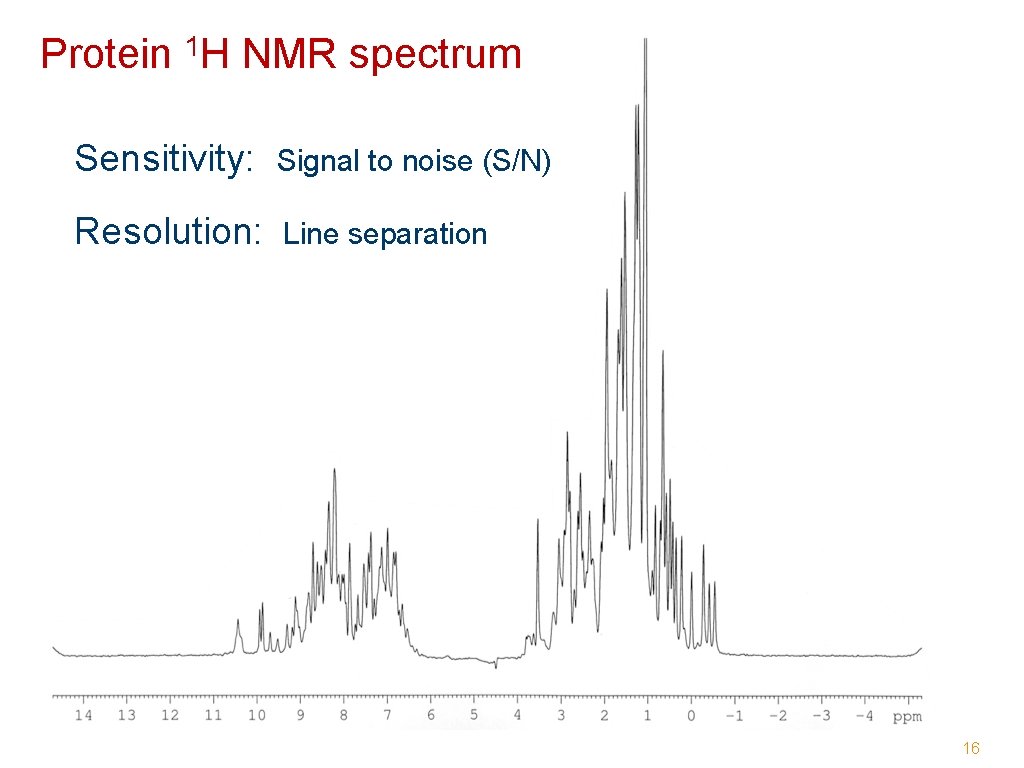
Protein 1 H NMR spectrum Sensitivity: Signal to noise (S/N) Resolution: Line separation 16
Why multidimensional NMR • multidimensional NMR experiments - resolve overlapping signals ➡ enables assignment of all signals - encode structural or dynamical information ➡ enables structure determination ➡ enables study of dynamics 17
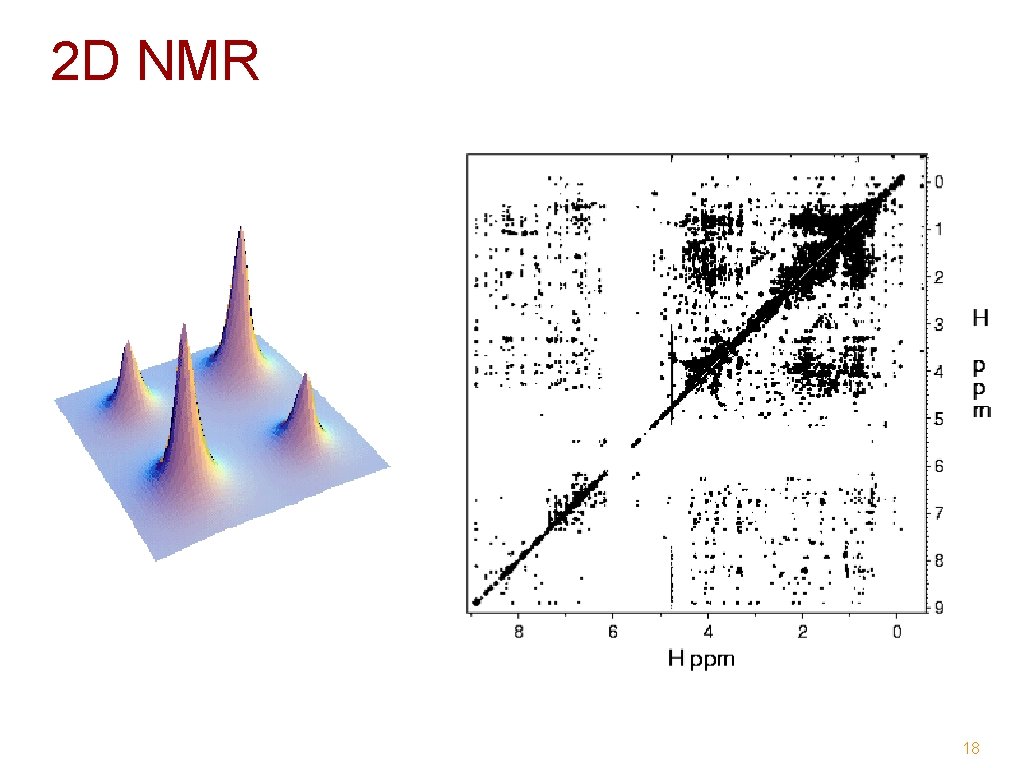
2 D NMR 18
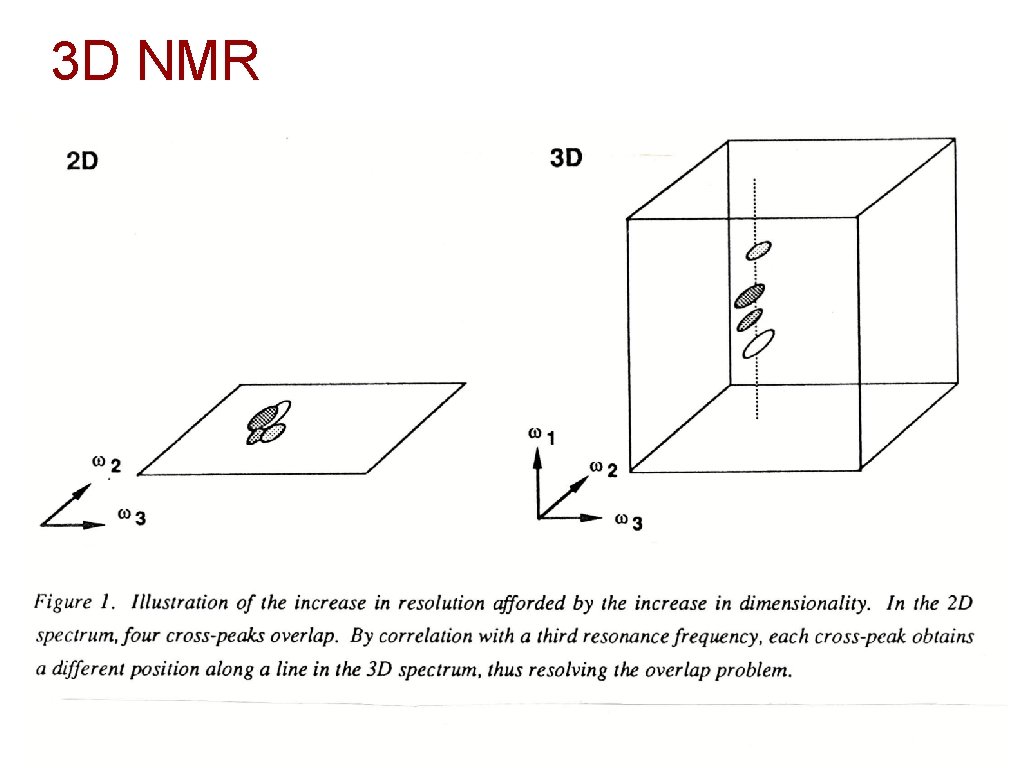
3 D NMR 19
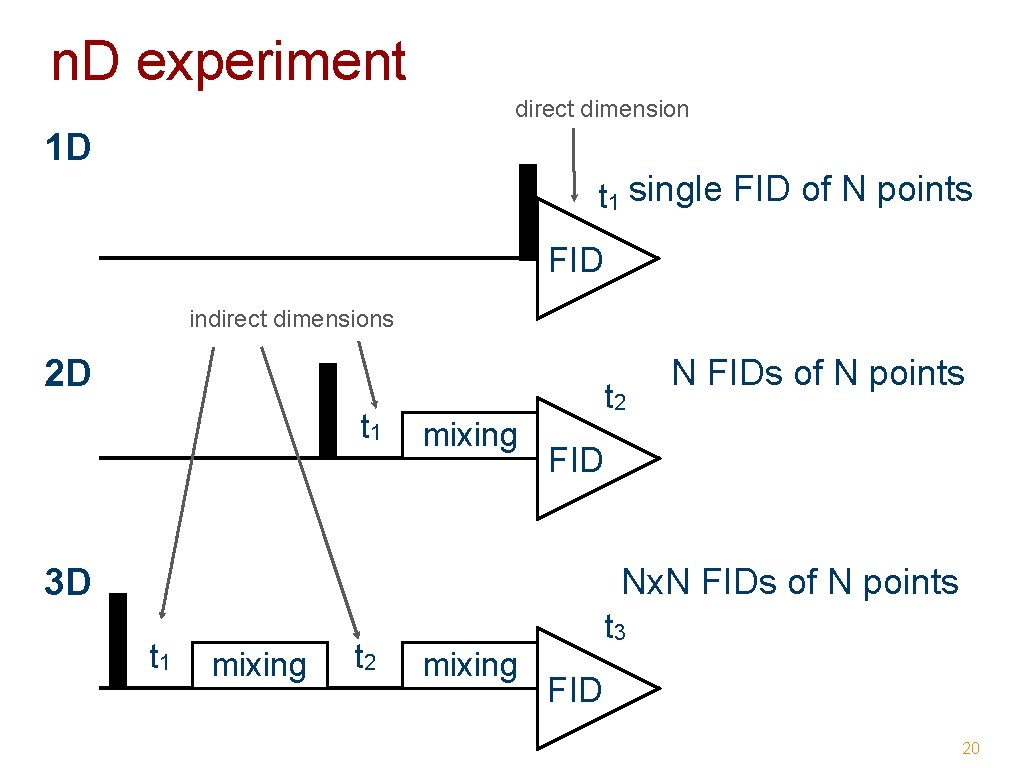
n. D experiment direct dimension 1 D t 1 single FID of N points FID indirect dimensions 2 D t 1 t 2 mixing N FIDs of N points FID 3 D Nx. N FIDs of N points t 1 mixing t 2 t 3 mixing FID 20
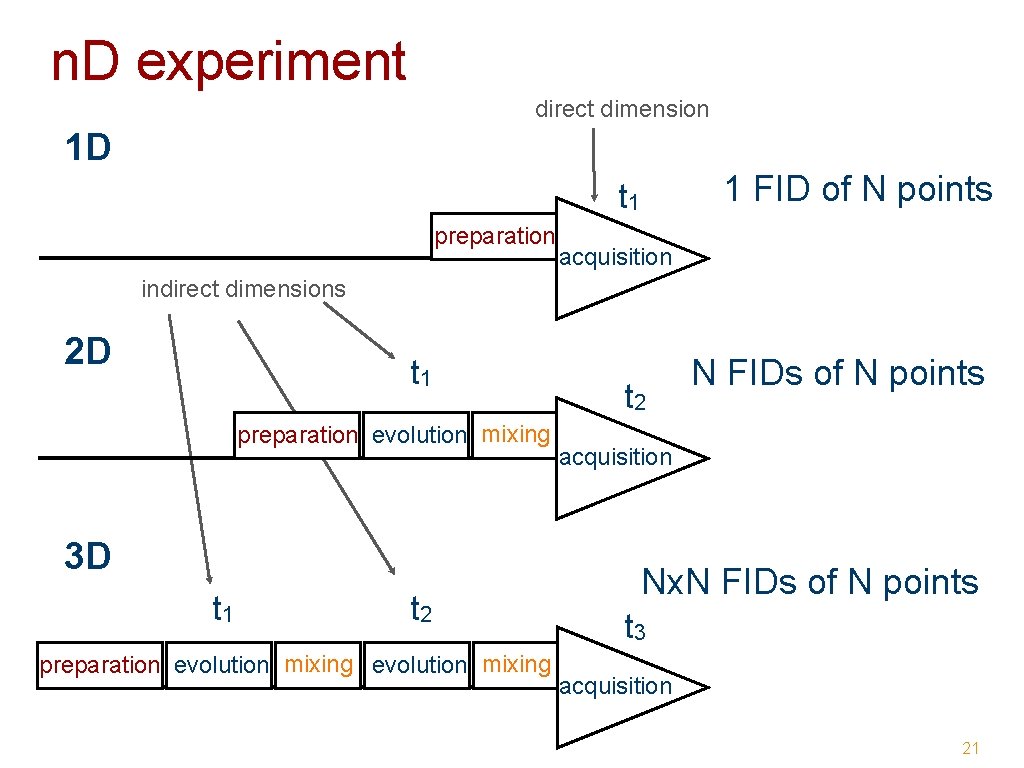
n. D experiment direct dimension 1 D 1 FID of N points t 1 preparation acquisition indirect dimensions 2 D t 1 preparation evolution mixing 3 D t 1 t 2 preparation evolution mixing t 2 N FIDs of N points acquisition Nx. N FIDs of N points t 3 acquisition 21
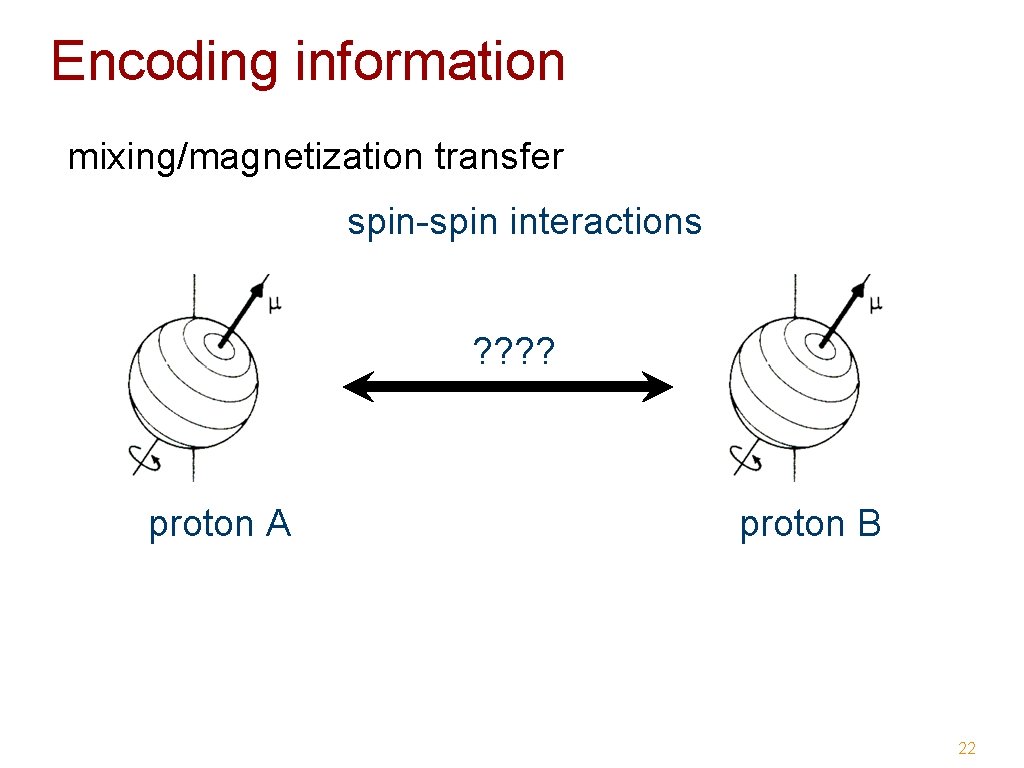
Encoding information mixing/magnetization transfer spin-spin interactions ? ? proton A proton B 22
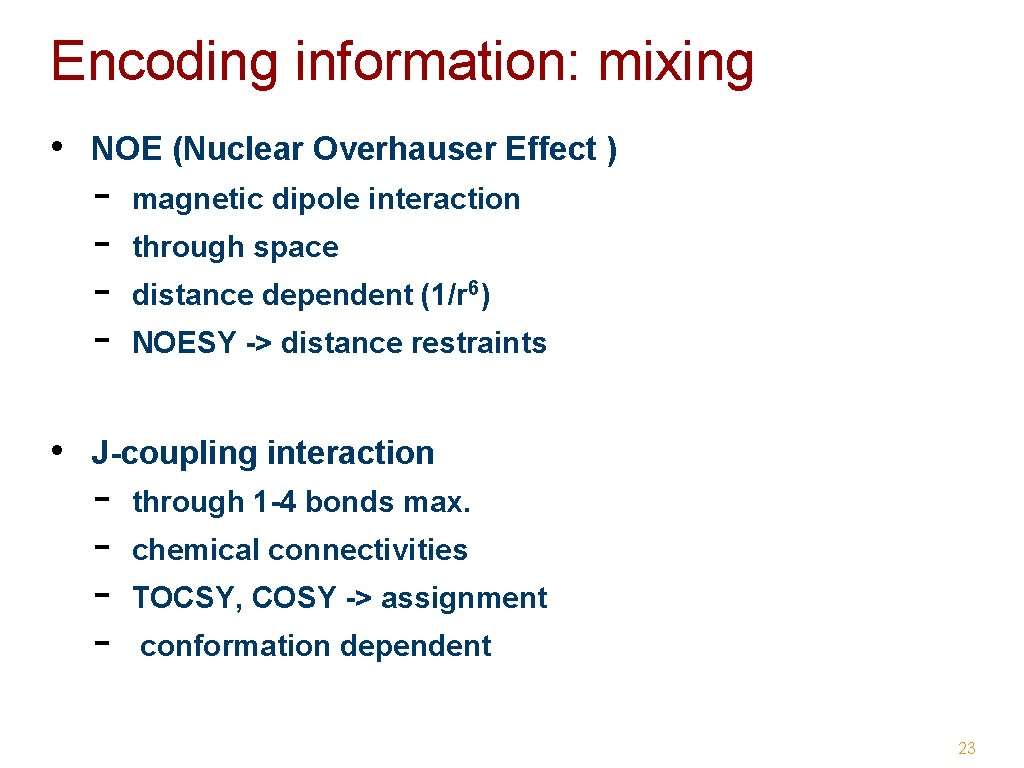
Encoding information: mixing • NOE (Nuclear Overhauser Effect ) - magnetic dipole interaction through space distance dependent (1/r 6) NOESY -> distance restraints • J-coupling interaction - through 1 -4 bonds max. chemical connectivities TOCSY, COSY -> assignment conformation dependent 23
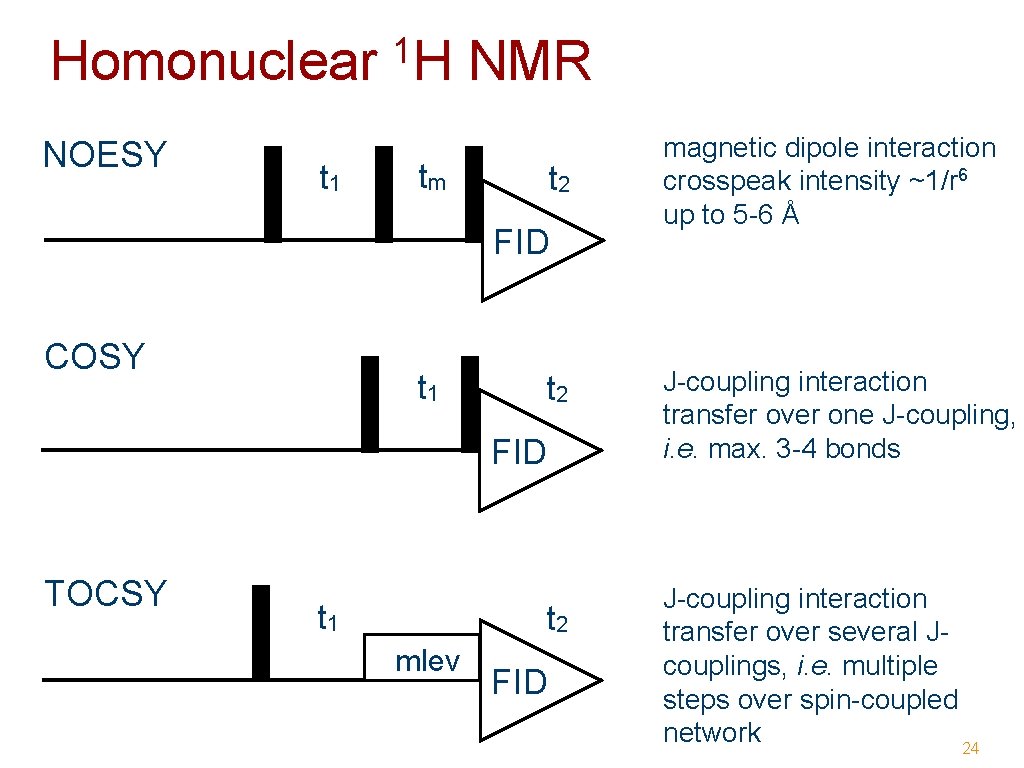
Homonuclear NOESY t 1 1 H NMR tm t 2 FID COSY t 1 t 2 FID TOCSY t 1 t 2 mlev FID magnetic dipole interaction crosspeak intensity ~1/r 6 up to 5 -6 Å J-coupling interaction transfer over one J-coupling, i. e. max. 3 -4 bonds J-coupling interaction transfer over several Jcouplings, i. e. multiple steps over spin-coupled network 24
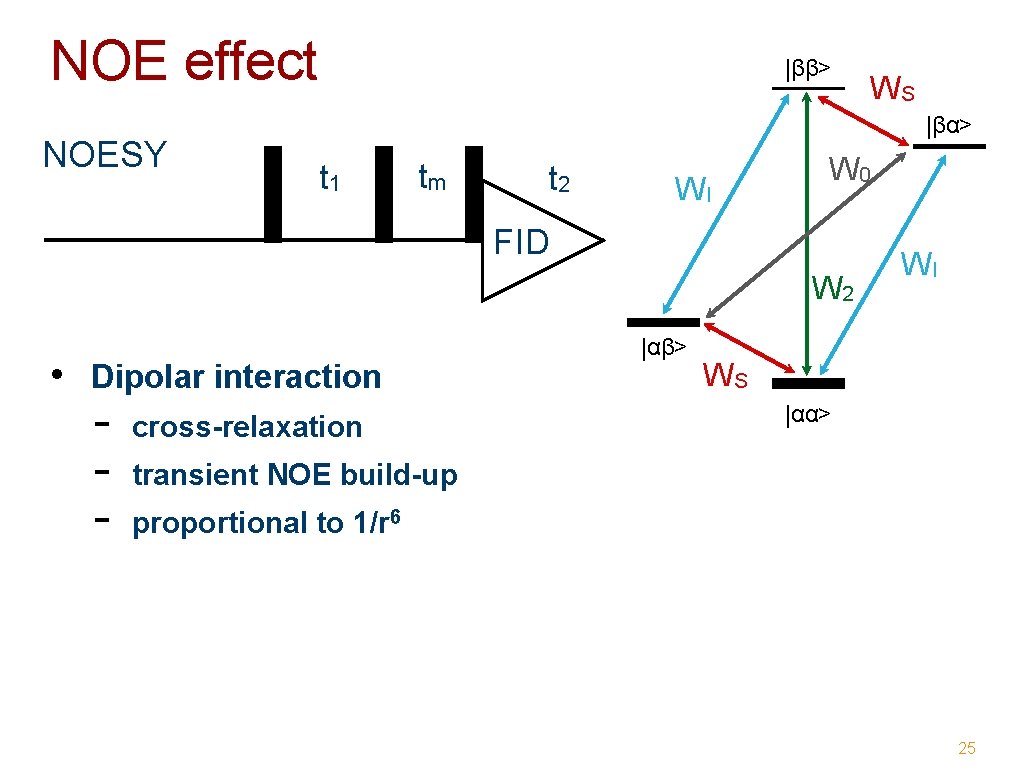
NOE effect NOESY |ββ> |βα> t 1 tm t 2 WI W 0 FID W 2 • Dipolar interaction - WS cross-relaxation |αβ> WI WS |αα> transient NOE build-up proportional to 1/r 6 25
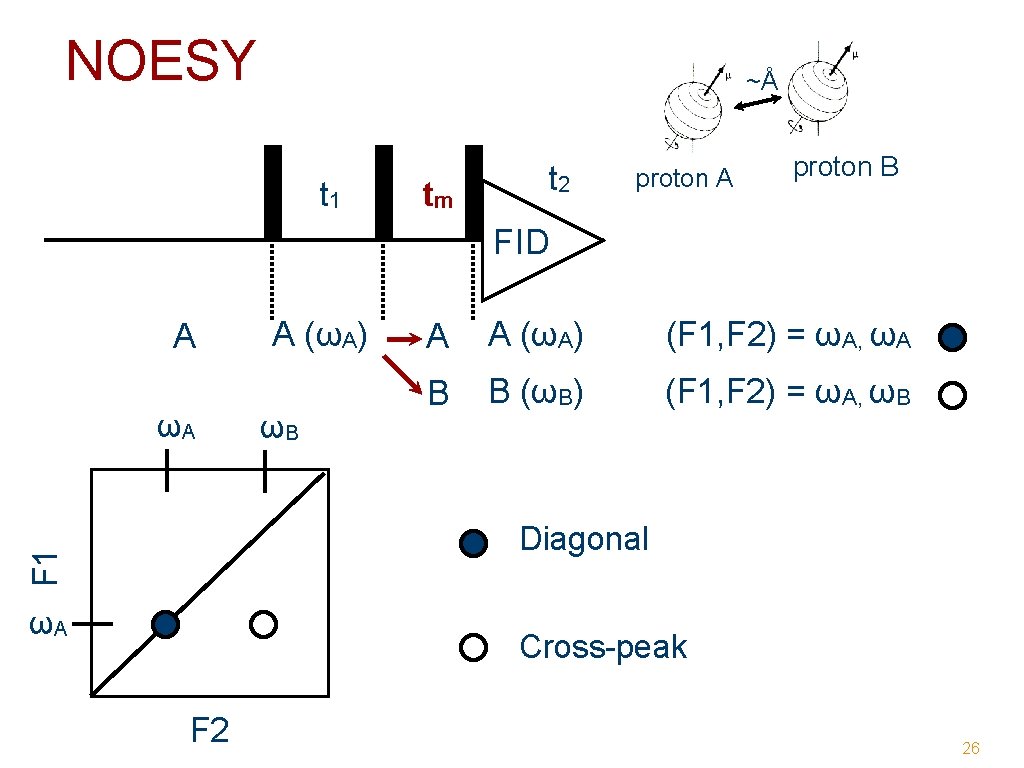
NOESY ~Å t 1 tm t 2 proton A proton B FID A ωA A (ωA) ωB A A (ωA) (F 1, F 2) = ωA, ωA B B (ωB) (F 1, F 2) = ωA, ωB F 1 Diagonal ωA Cross-peak F 2 26
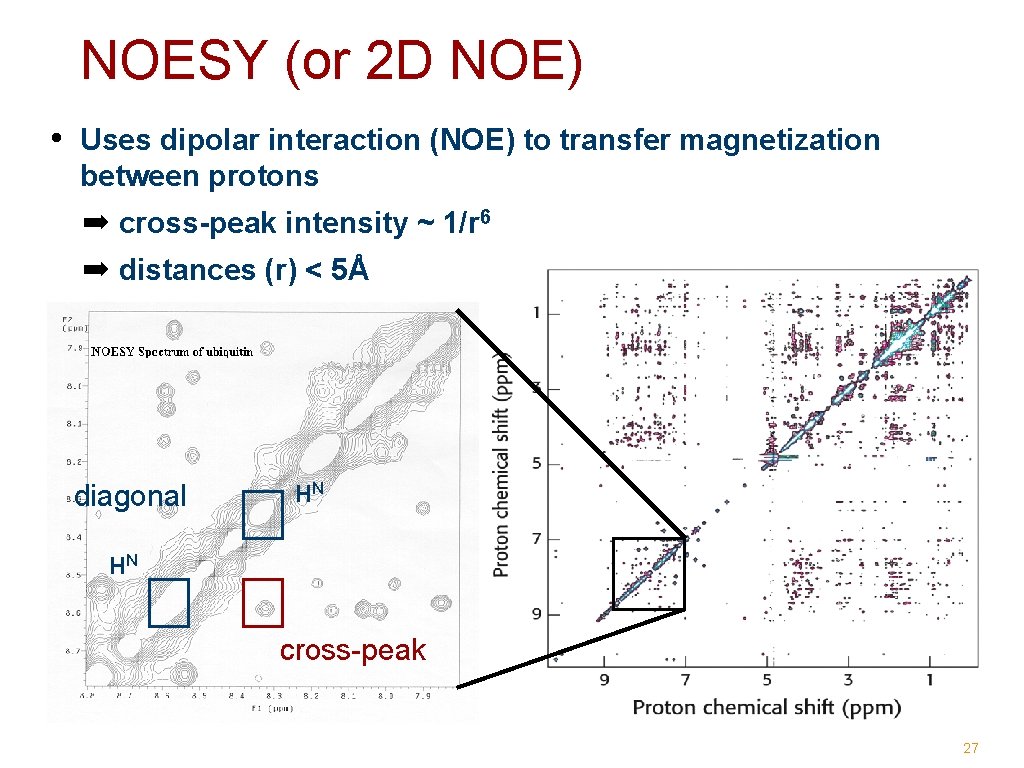
NOESY (or 2 D NOE) • Uses dipolar interaction (NOE) to transfer magnetization between protons ➡ cross-peak intensity ~ 1/r 6 ➡ distances (r) < 5Å diagonal HN HN cross-peak 27
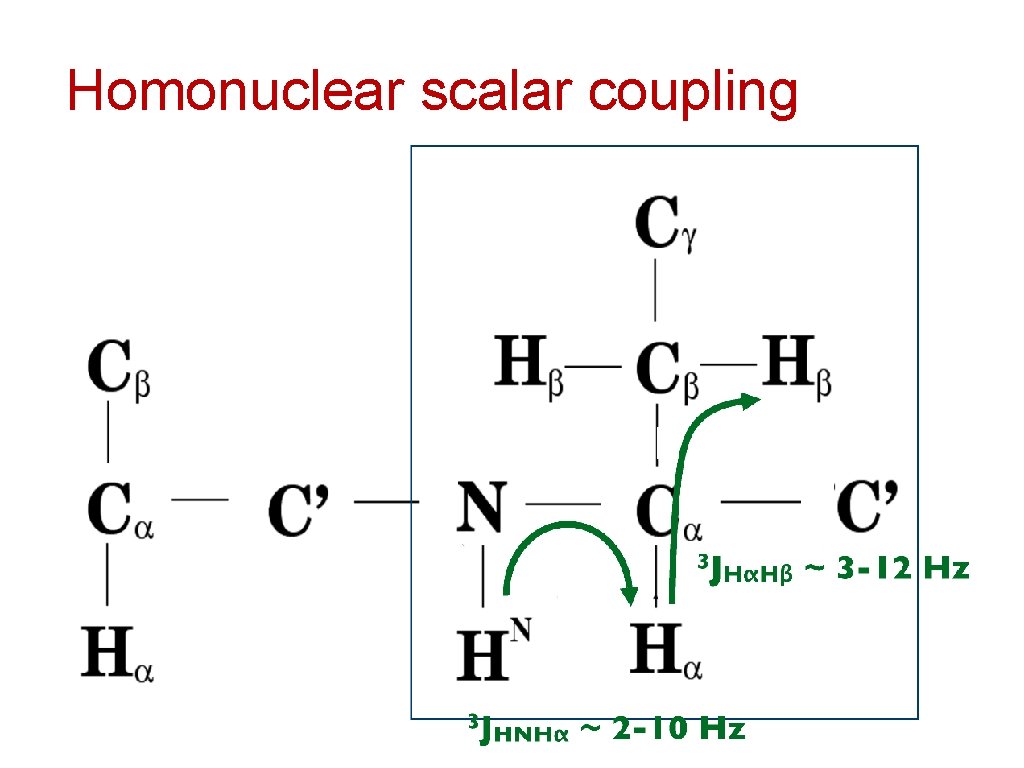
Homonuclear scalar coupling
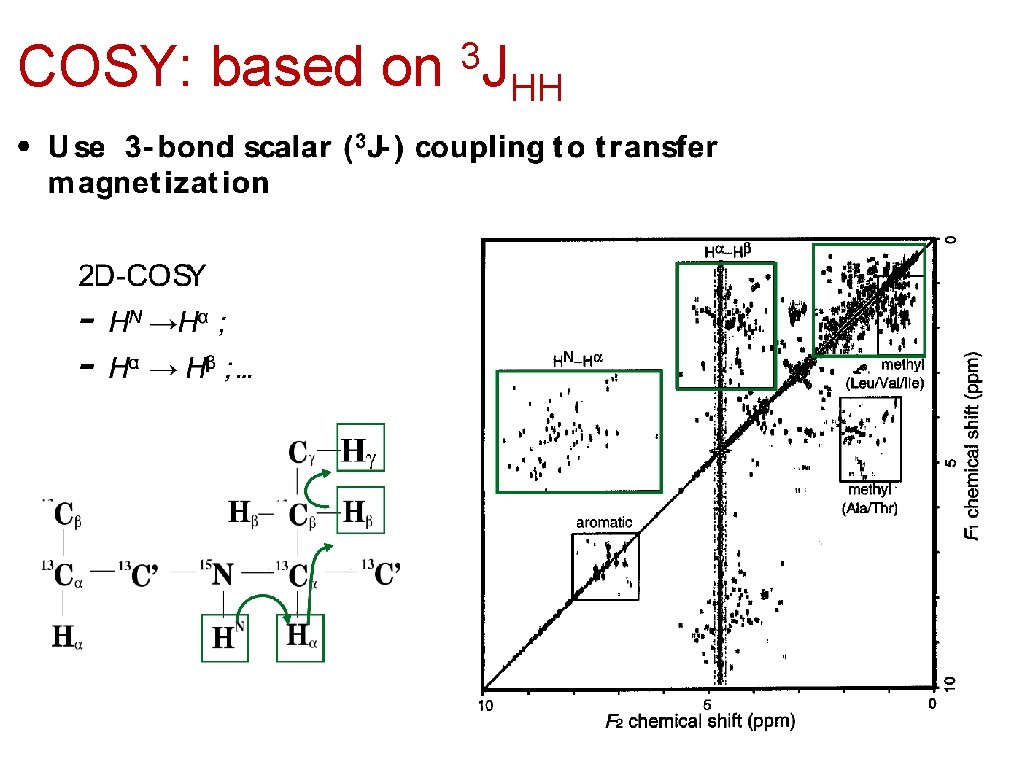
COSY: based on 3 J HH
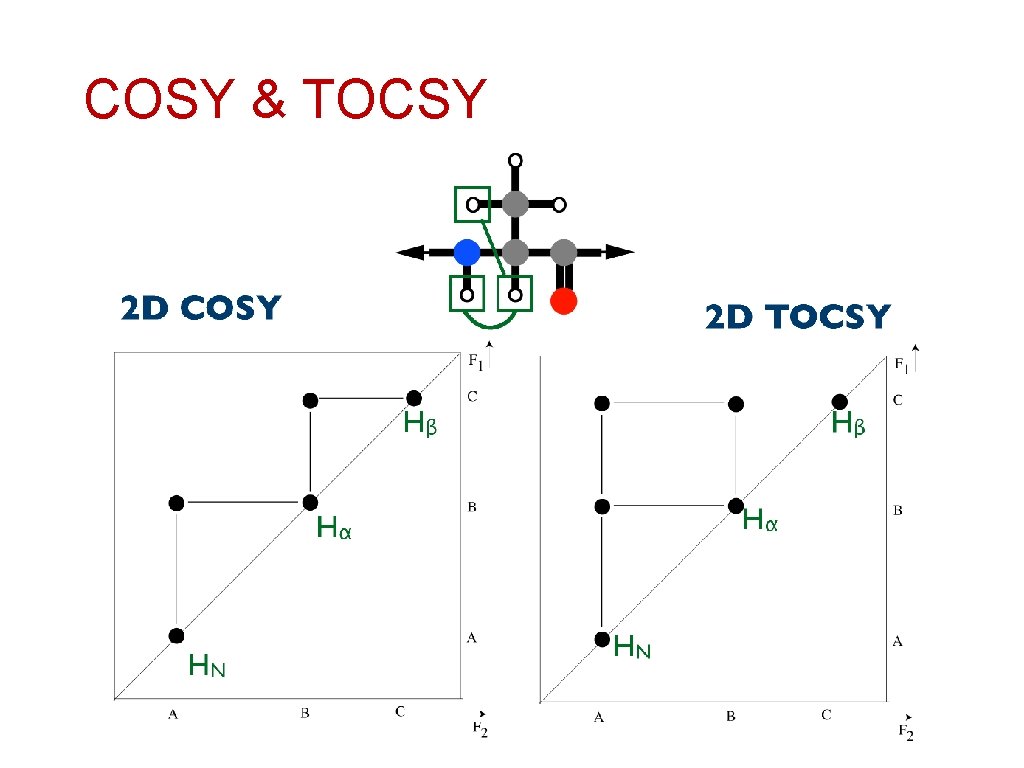
COSY & TOCSY

COSY & TOCSY COSY o TOCSY *
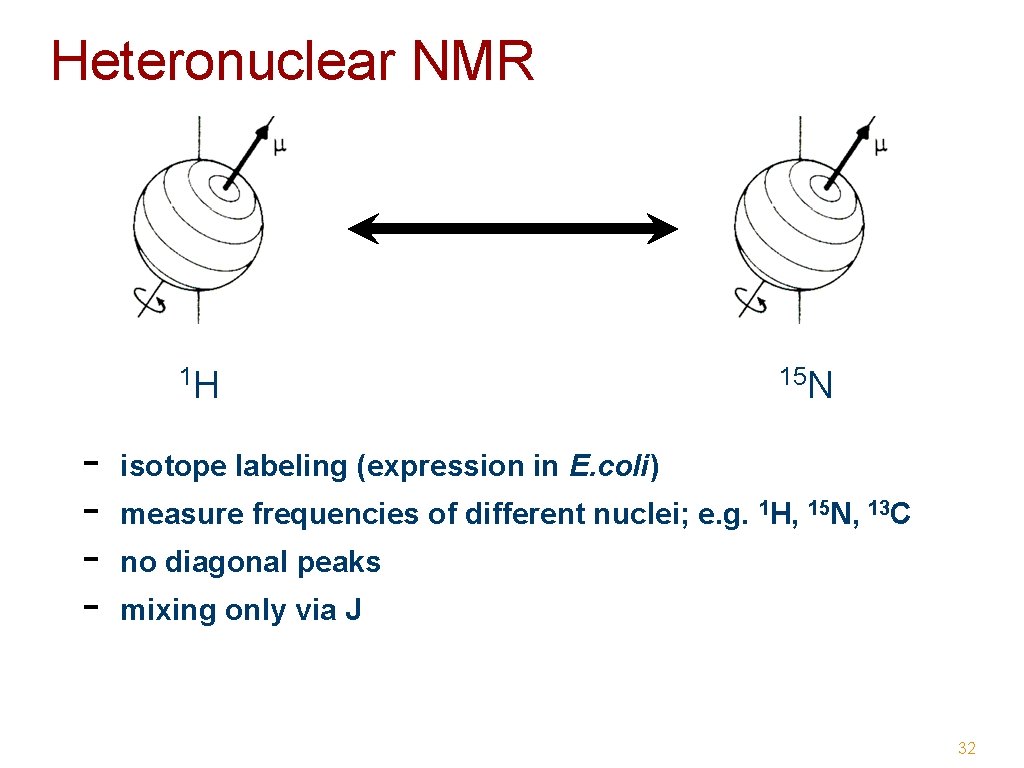
Heteronuclear NMR 1 H - 15 N isotope labeling (expression in E. coli) measure frequencies of different nuclei; e. g. 1 H, 15 N, 13 C no diagonal peaks mixing only via J 32
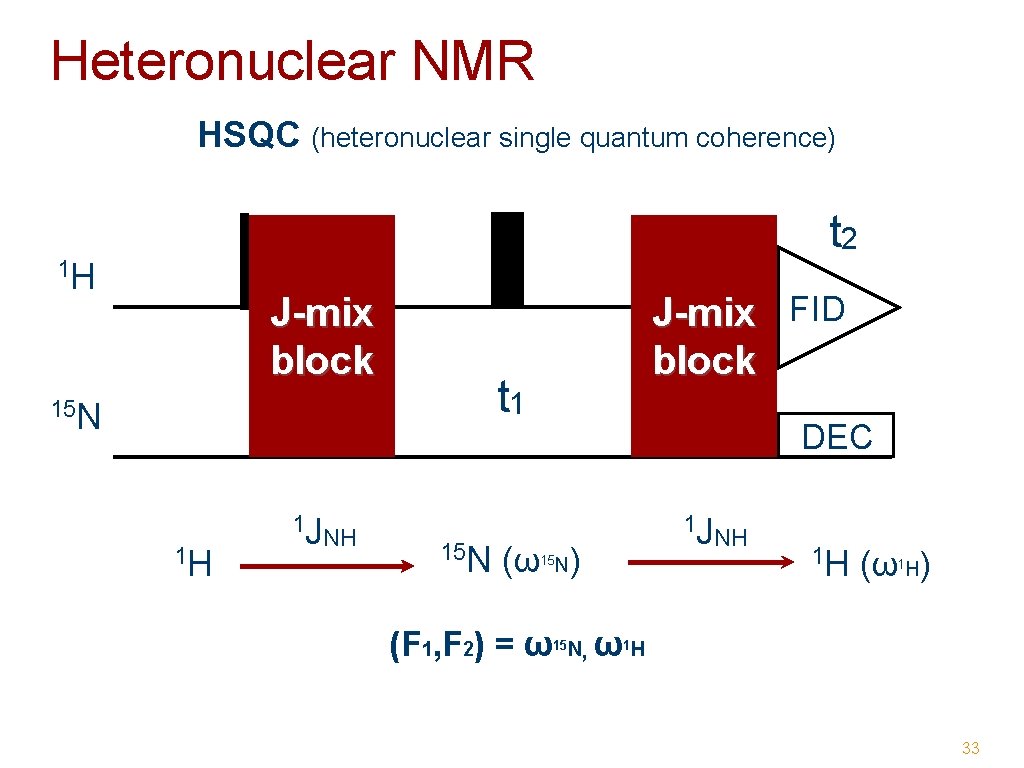
Heteronuclear NMR HSQC (heteronuclear single quantum coherence) t 2 1 H J-mix block t 1 15 N 1 H 1 J NH J-mix FID block 15 N DEC 1 J NH ( ω N) 15 (F 1, F 2) = ω 15 N 1 H ( ω H) 1 , ω1 H 33
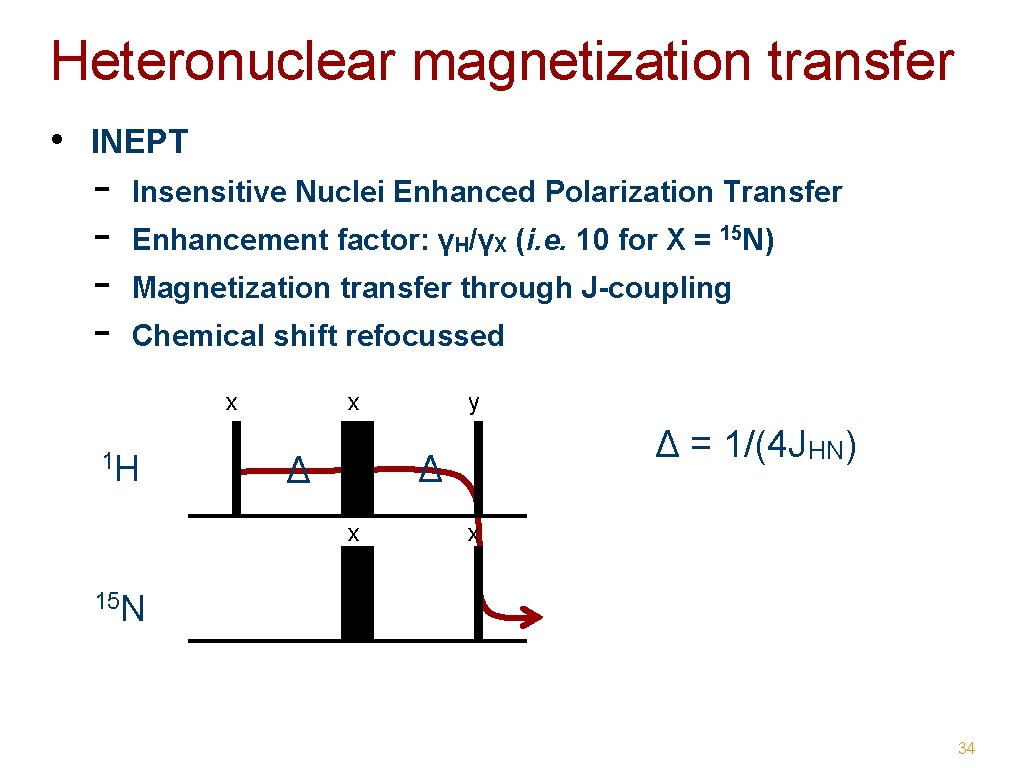
Heteronuclear magnetization transfer • INEPT - Insensitive Nuclei Enhanced Polarization Transfer Enhancement factor: γH/γX (i. e. 10 for X = 15 N) Magnetization transfer through J-coupling Chemical shift refocussed x 1 H x y Δ = 1/(4 JHN) Δ Δ x x 15 N 34
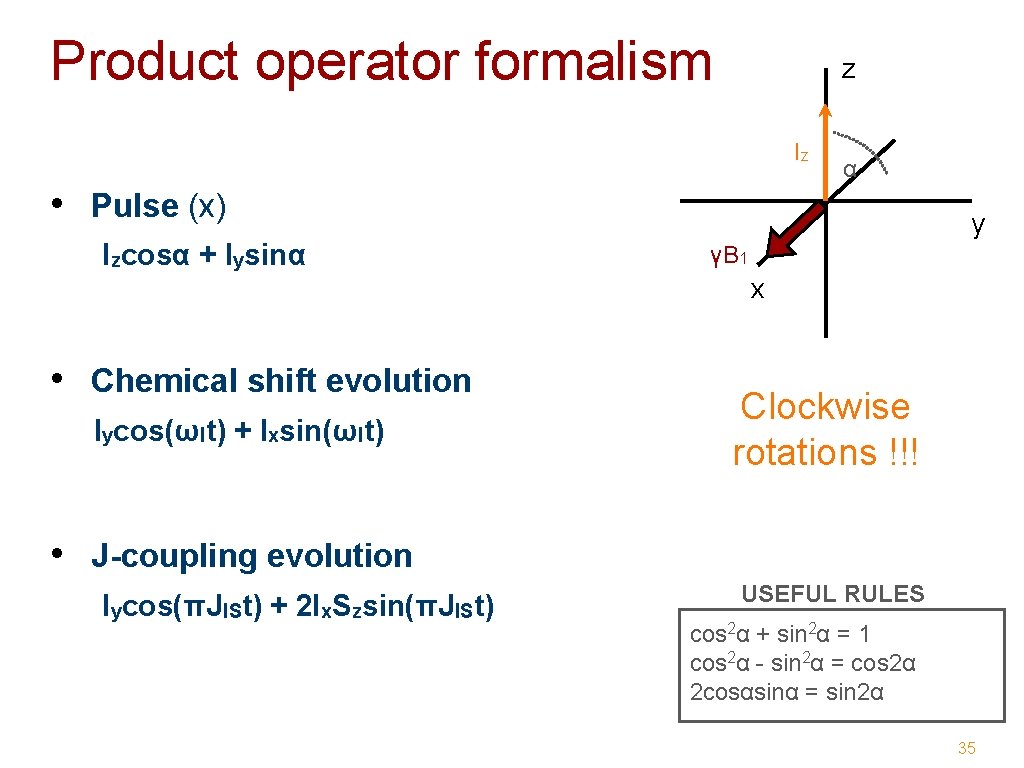
Product operator formalism z Iz • Pulse (x) Izcosα + Iysinα α y γB 1 x • Chemical shift evolution Iycos(ωIt) + Ixsin(ωIt) Clockwise rotations !!! • J-coupling evolution Iycos(πJISt) + 2 Ix. Szsin(πJISt) USEFUL RULES cos 2α + sin 2α = 1 cos 2α - sin 2α = cos 2α 2 cosαsinα = sin 2α 35
15 N-HSQC 36
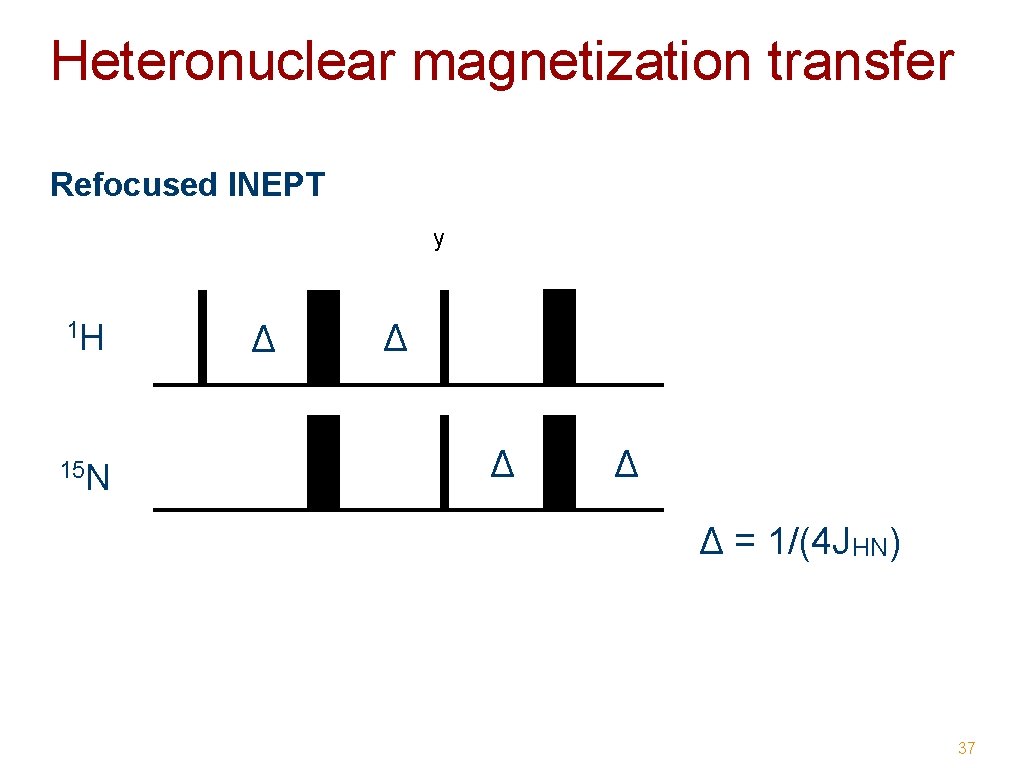
Heteronuclear magnetization transfer Refocused INEPT y 1 H 15 N Δ Δ Δ = 1/(4 JHN) 37
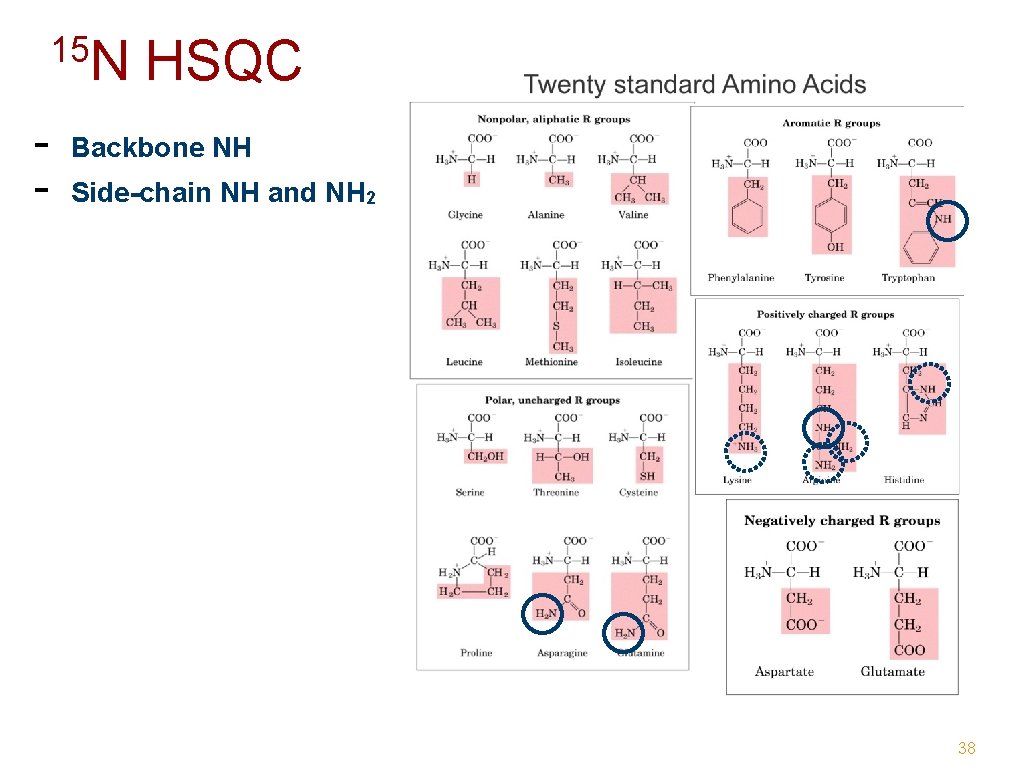
15 N - HSQC Backbone NH Side-chain NH and NH 2 38
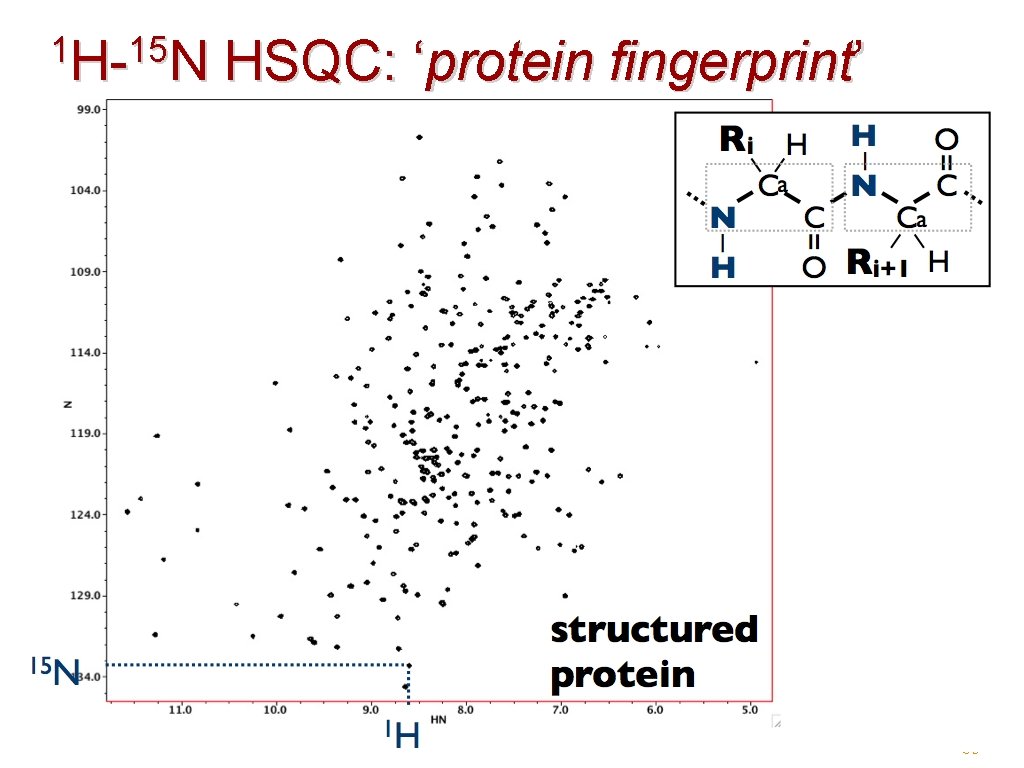
1 H-15 N H- N HSQC: ‘protein fingerprint’ 39
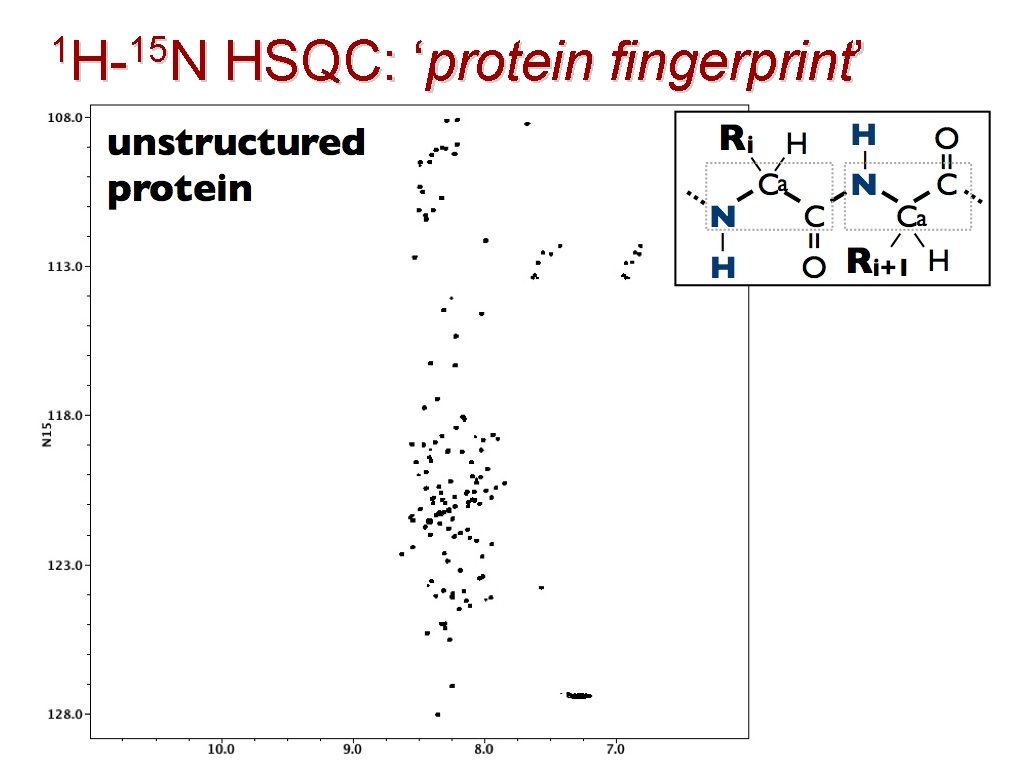
1 H-15 N H- N HSQC: ‘protein fingerprint’ 40
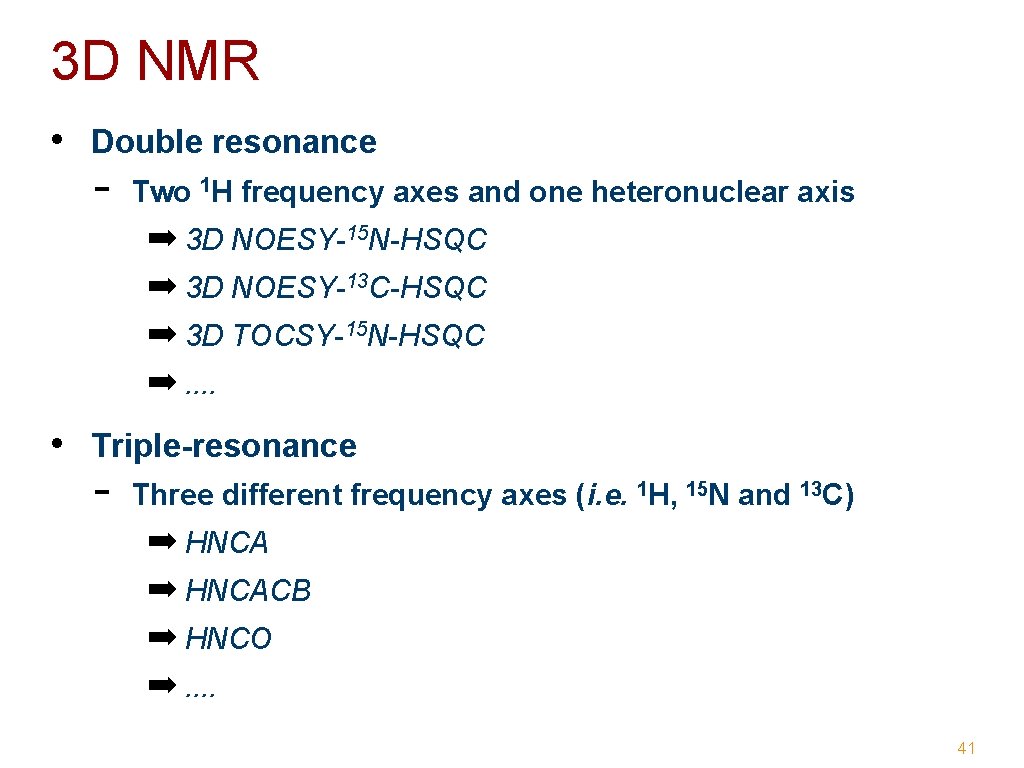
3 D NMR • Double resonance - Two 1 H frequency axes and one heteronuclear axis ➡ 3 D NOESY-15 N-HSQC ➡ 3 D NOESY-13 C-HSQC ➡ 3 D TOCSY-15 N-HSQC ➡. . • Triple-resonance - Three different frequency axes (i. e. 1 H, 15 N and 13 C) ➡ HNCACB ➡ HNCO ➡. . 41
![3 D 1 15 TOCSY-[ H- N]-HSQC 3 HN atoms same δHN Same δHN 3 D 1 15 TOCSY-[ H- N]-HSQC 3 HN atoms same δHN Same δHN](http://slidetodoc.com/presentation_image_h/af0501cb1dc4822f7de293a952247998/image-42.jpg)
3 D 1 15 TOCSY-[ H- N]-HSQC 3 HN atoms same δHN Same δHN different δ 15 N 1 H 1 H N TOCSY HSQC 1 H → 1 HN → 15 N → 1 HN t 1 t 2 t 3 15 N FT F 2 F 3 F 1 42
![3 D 1 15 TOCSY-[ H- N]-HSQC strip 1 H 1 H N 15 3 D 1 15 TOCSY-[ H- N]-HSQC strip 1 H 1 H N 15](http://slidetodoc.com/presentation_image_h/af0501cb1dc4822f7de293a952247998/image-43.jpg)
3 D 1 15 TOCSY-[ H- N]-HSQC strip 1 H 1 H N 15 N [1 H, 15 N]-HSQC projection 43
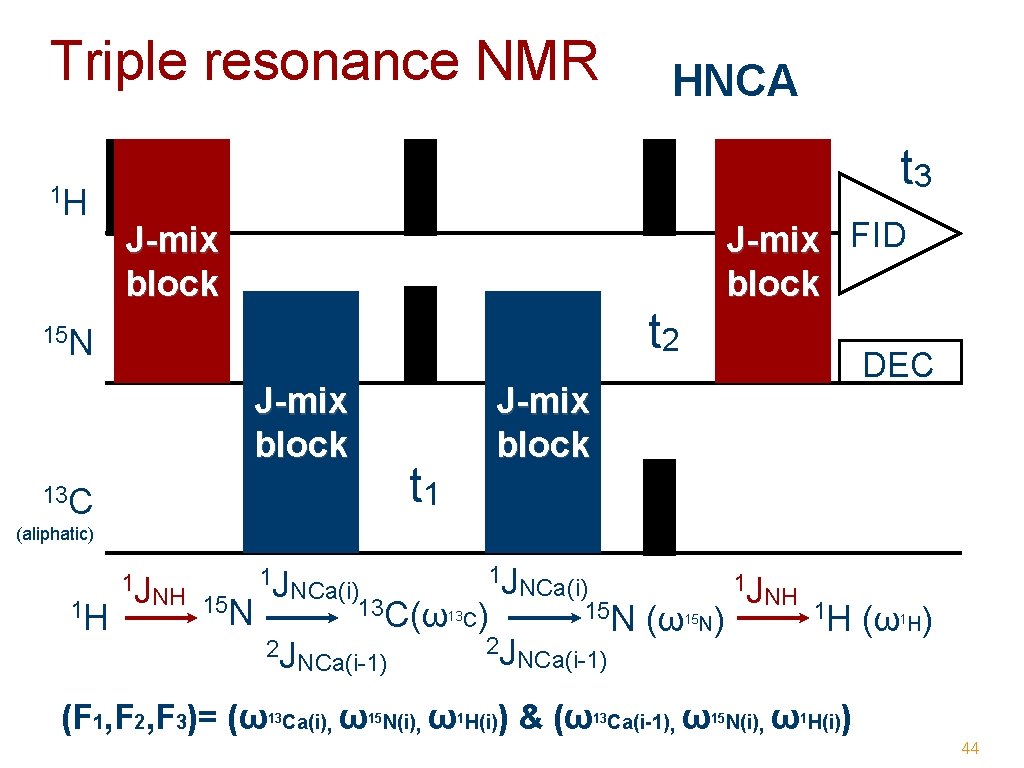
Triple resonance NMR 1 H HNCA t 3 J-mix FID block J-mix block t 2 15 N J-mix block 13 C t 1 DEC J-mix block (aliphatic) 1 H 1 J NCa(i) 15 N C(ω13 C) 2 J 2 J NCa(i-1) 1 J 1 J NCa(i) NH 15 13 N (F 1, F 2, F 3)= (ω 13 Ca(i) , ω15 N(i), ω1 H(i)) & (ω ( ω N) 13 Ca(i-1) 15 1 J NH 1 H ( ω H) 1 , ω15 N(i), ω1 H(i)) 44
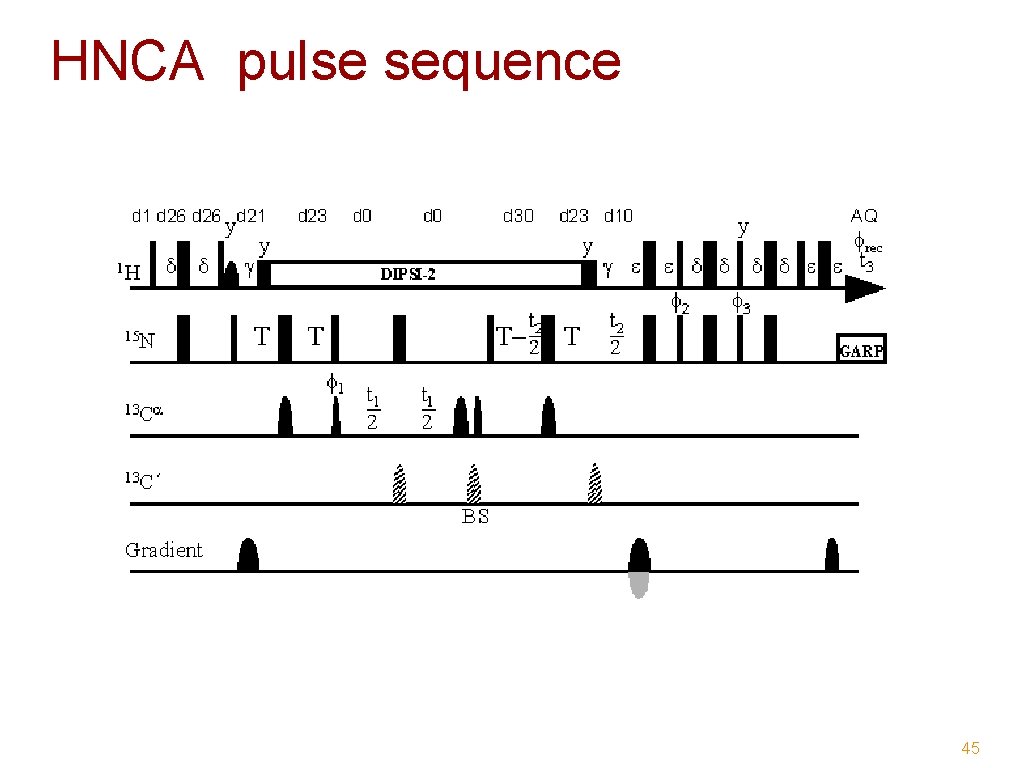
HNCA pulse sequence 45
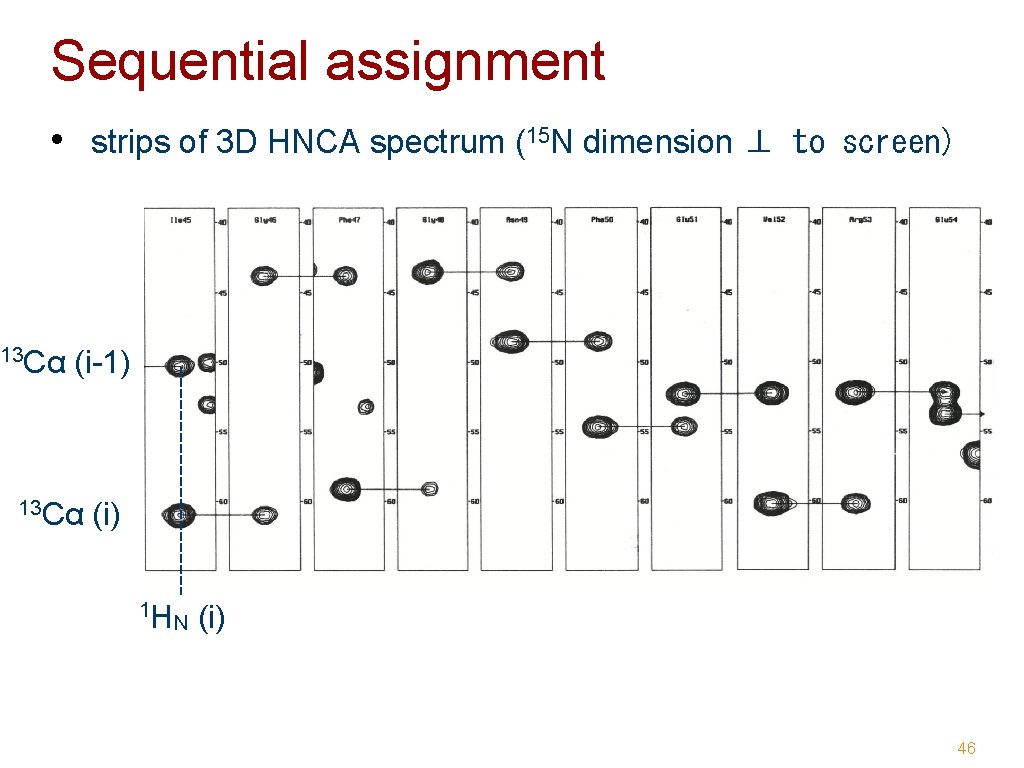
Sequential assignment • strips of 3 D HNCA spectrum (15 N dimension ⊥ to screen) 13 Cα (i-1) 13 Cα (i) 1 H N (i) 46
Key concepts multidimensional NMR • resolve overlapping signals • mixing/magnetization transfer • NOESY, TOCSY, COSY • HSQC • 3 D double resonance (3 D NOESY-HSQC, 3 D TOCSY-HSQC) • 3 D triple resonance (e. g. HNCA) 47
- Slides: 47