Biology Primer Basic principles OrganismsCells as Basic Units
Biology Primer • • • Basic principles Organisms/Cells as Basic Units Biochemical Components of Cells Genetic processes Fundamental Molecular Genetics BIO 520 Bioinformatics Jim Lund
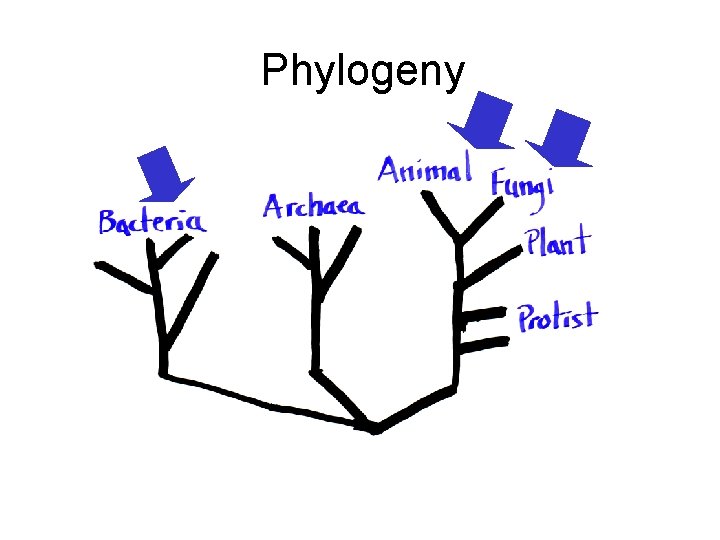
Phylogeny
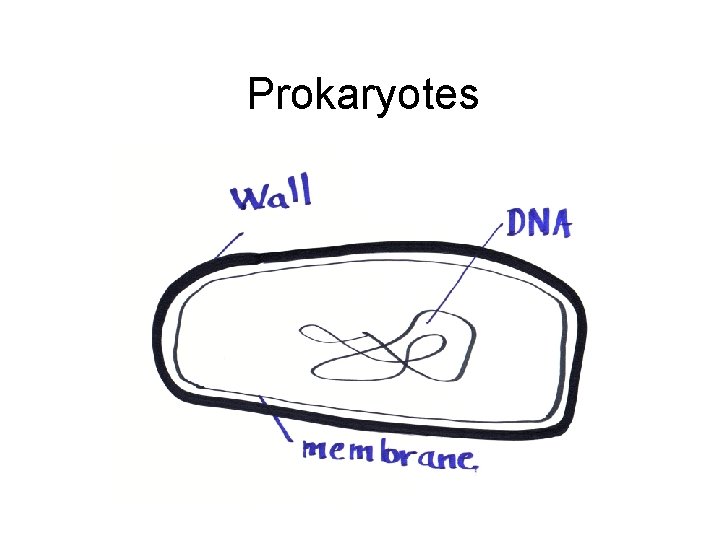
Prokaryotes
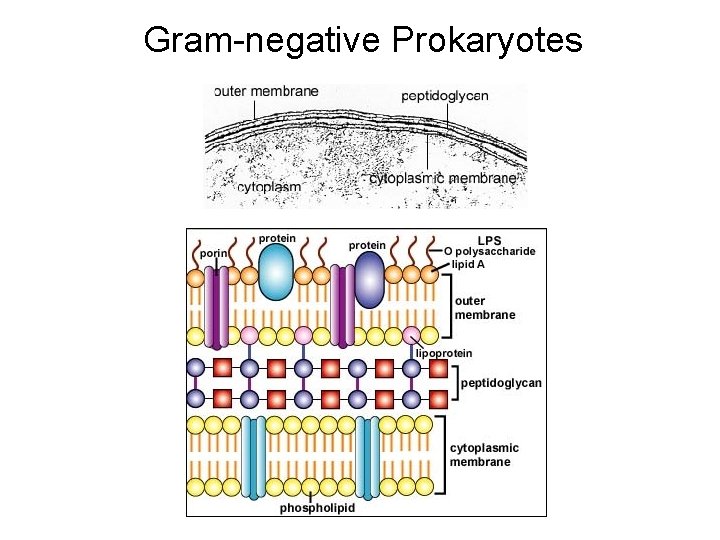
Gram-negative Prokaryotes
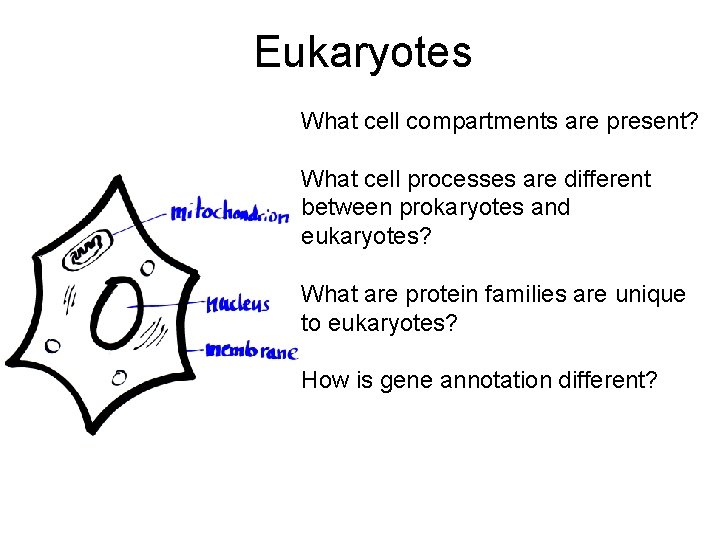
Eukaryotes What cell compartments are present? What cell processes are different between prokaryotes and eukaryotes? What are protein families are unique to eukaryotes? How is gene annotation different?
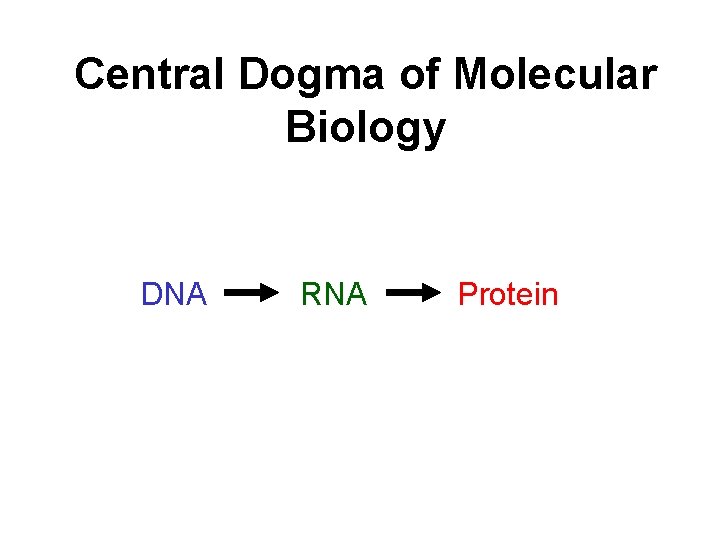
Central Dogma of Molecular Biology DNA RNA Protein
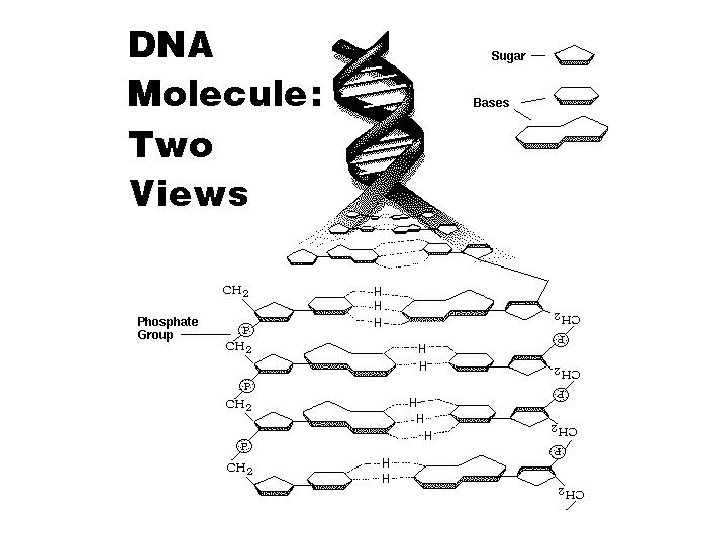
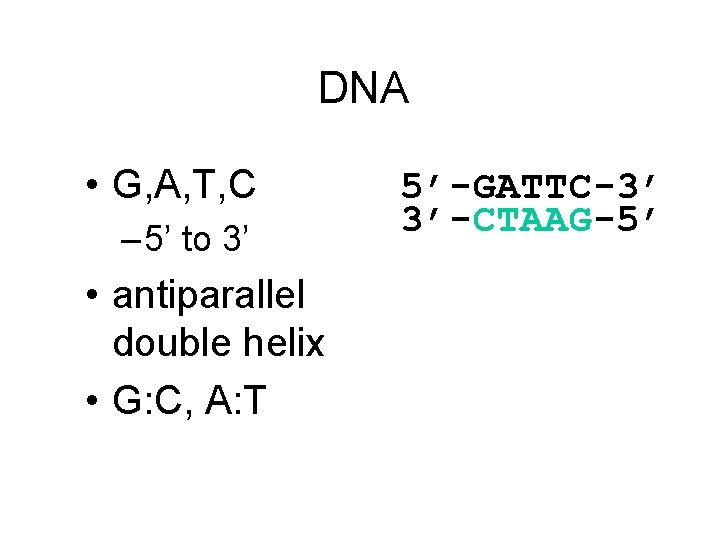
DNA • G, A, T, C – 5’ to 3’ • antiparallel double helix • G: C, A: T 5’-GATTC-3’ 3’-CTAAG-5’
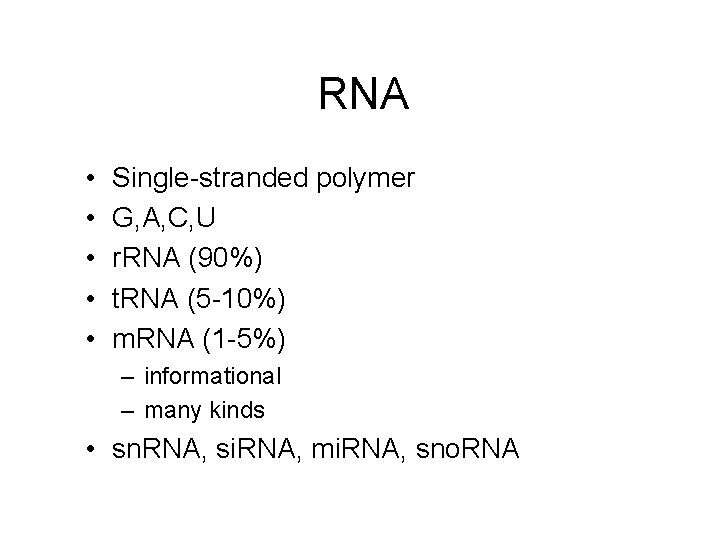
RNA • • • Single-stranded polymer G, A, C, U r. RNA (90%) t. RNA (5 -10%) m. RNA (1 -5%) – informational – many kinds • sn. RNA, si. RNA, mi. RNA, sno. RNA
Other Molecules • Lipids • Carbohydrates • Small molecules –Precursors, metabolites…
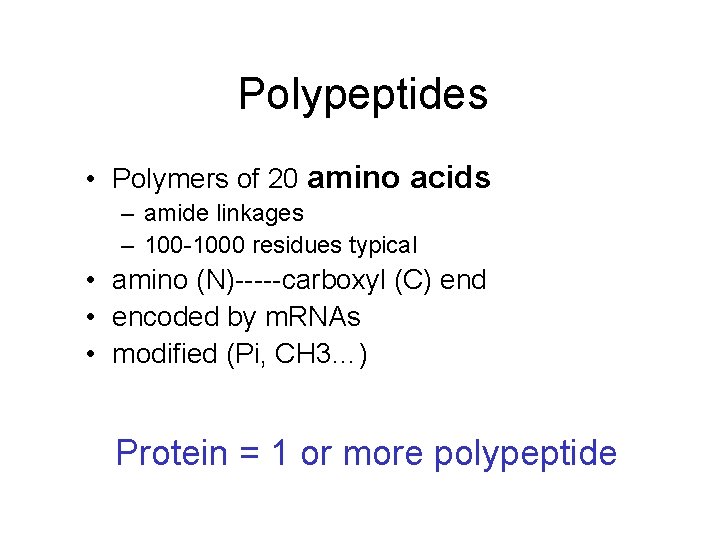
Polypeptides • Polymers of 20 amino acids – amide linkages – 100 -1000 residues typical • amino (N)-----carboxyl (C) end • encoded by m. RNAs • modified (Pi, CH 3…) Protein = 1 or more polypeptide
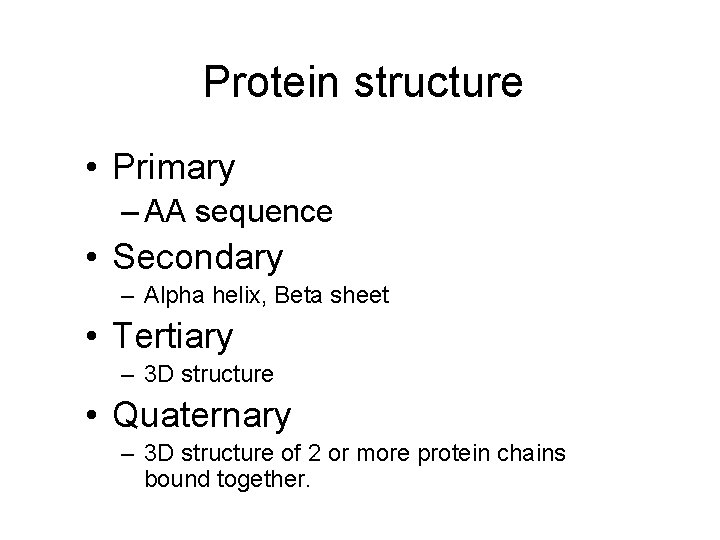
Protein structure • Primary – AA sequence • Secondary – Alpha helix, Beta sheet • Tertiary – 3 D structure • Quaternary – 3 D structure of 2 or more protein chains bound together.

“JOE AVERAGE” Polypeptide • ~350 amino acids – Rare vs common aa’s • Amino acids – MW=~110 Da
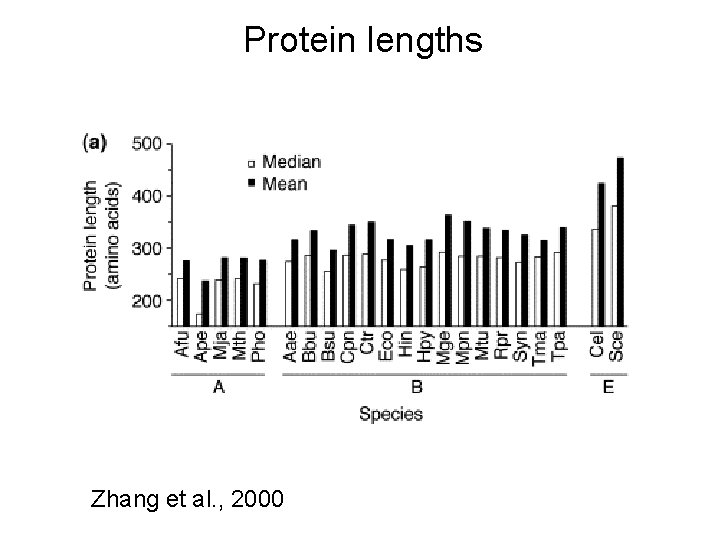
Protein lengths Zhang et al. , 2000
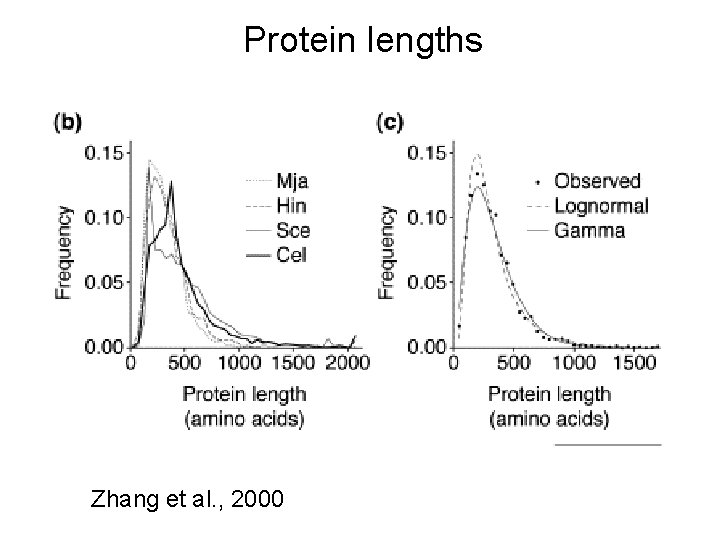
Protein lengths Zhang et al. , 2000
Proteins • • • Enzymes Receptors Transporters Structural components Regulatory factors
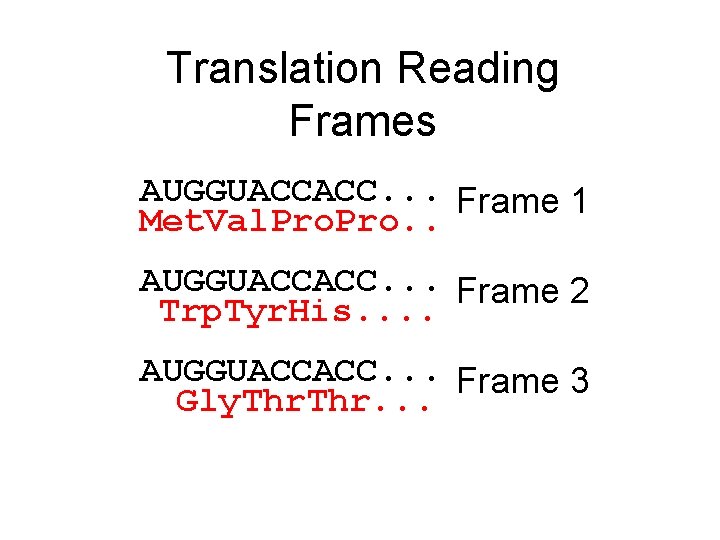
Translation Reading Frames AUGGUACCACC. . . Frame 1 Met. Val. Pro. . AUGGUACCACC. . . Frame 2 Trp. Tyr. His. . AUGGUACCACC. . . Frame 3 Gly. Thr. . .
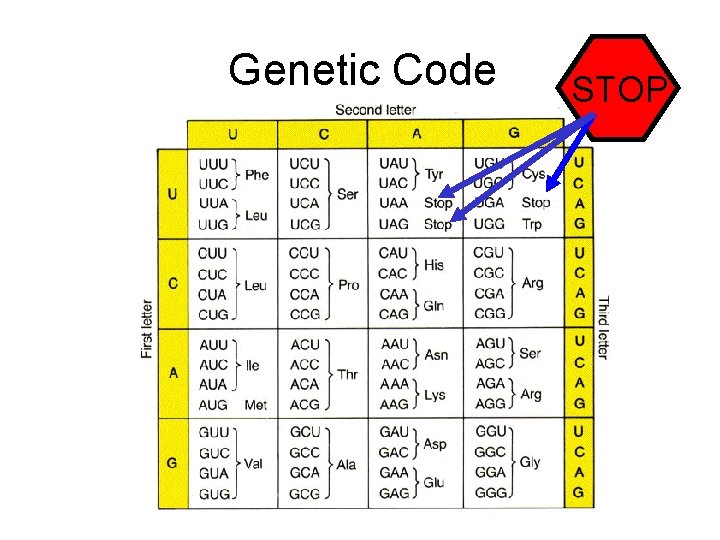
Genetic Code STOP
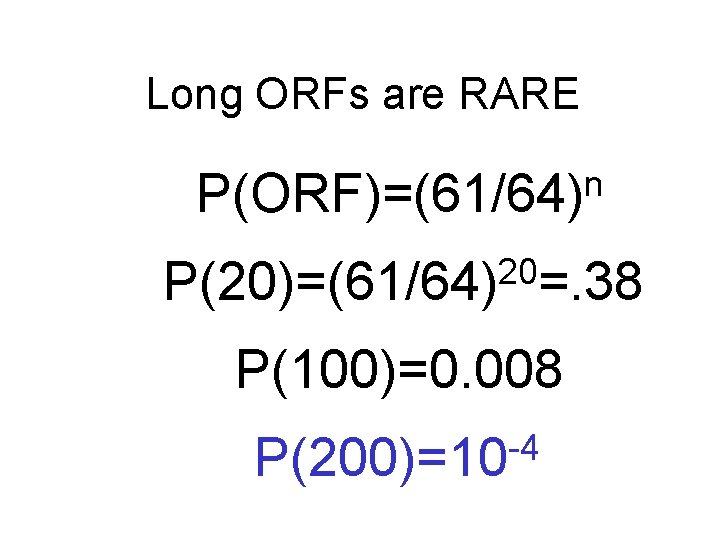
Long ORFs are RARE n P(ORF)=(61/64) 20 P(20)=(61/64) =. 38 P(100)=0. 008 -4 P(200)=10
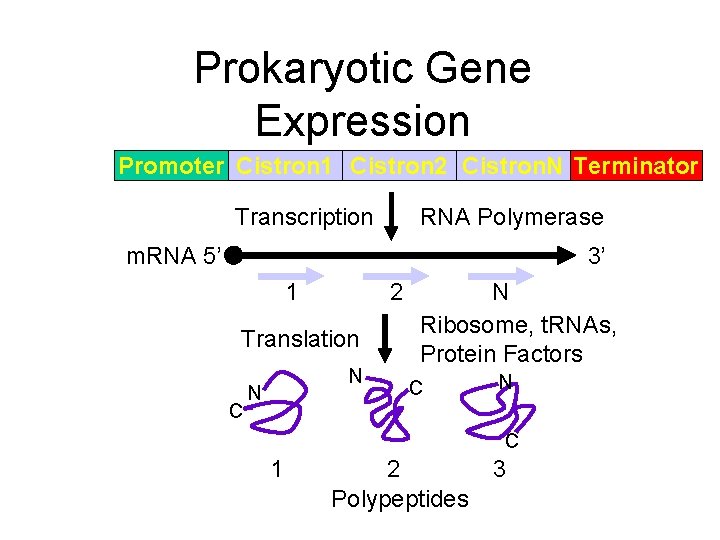
Prokaryotic Gene Expression Promoter Cistron 1 Cistron 2 Cistron. N Terminator Transcription RNA Polymerase m. RNA 5’ 3’ 1 2 Translation C N N N Ribosome, t. RNAs, Protein Factors C N C 1 2 Polypeptides 3
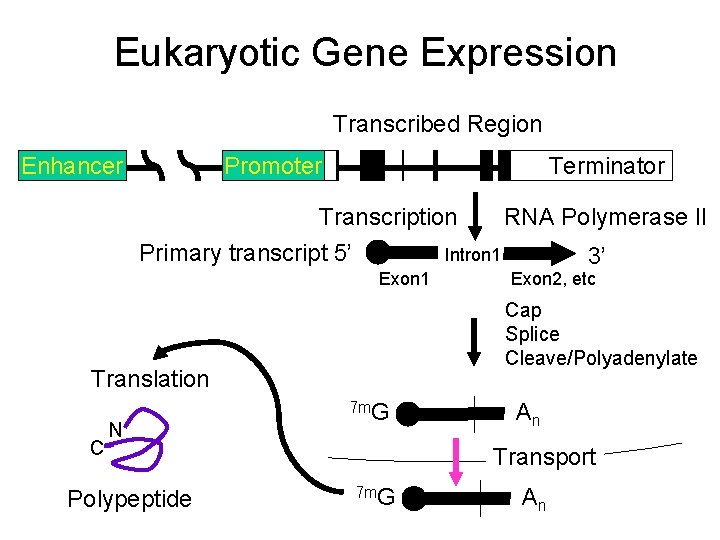
Eukaryotic Gene Expression Transcribed Region Enhancer Promoter Terminator Transcription RNA Polymerase II Primary transcript 5’ Intron 1 3’ Exon 1 Cap Splice Cleave/Polyadenylate Translation C N Polypeptide Exon 2, etc 7 m. G An Transport 7 m. G An

“Joe Average” Gene Encodes 350 amino acid product • Euk: – Exons and introns, typically ~5 -10 – Enhancer/promoter region • Prok: 1 cistron (part of operon)
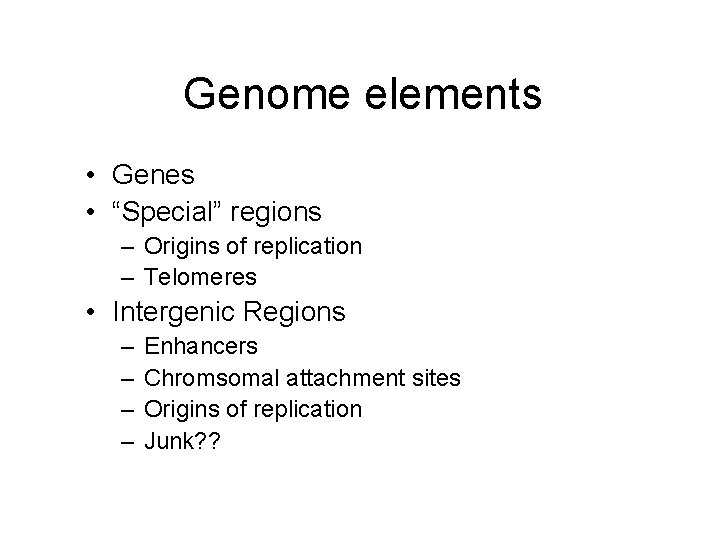
Genome elements • Genes • “Special” regions – Origins of replication – Telomeres • Intergenic Regions – – Enhancers Chromsomal attachment sites Origins of replication Junk? ?
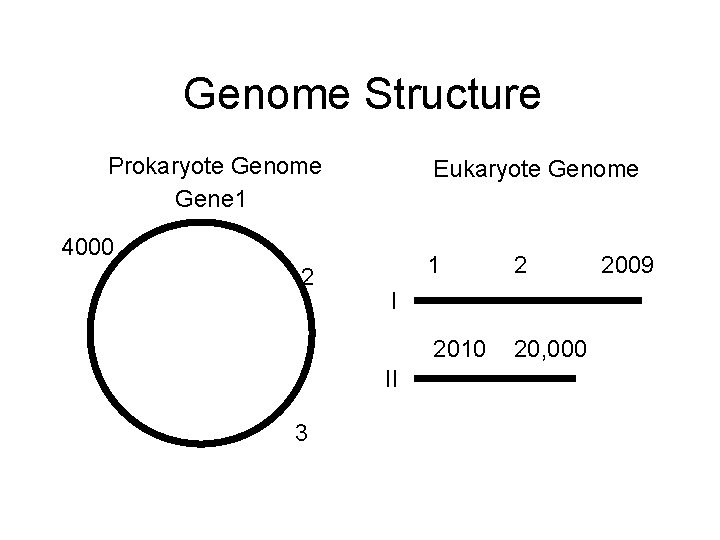
Genome Structure Prokaryote Genome Gene 1 Eukaryote Genome 4000 2 2 2010 20, 000 I II 3 1 2009
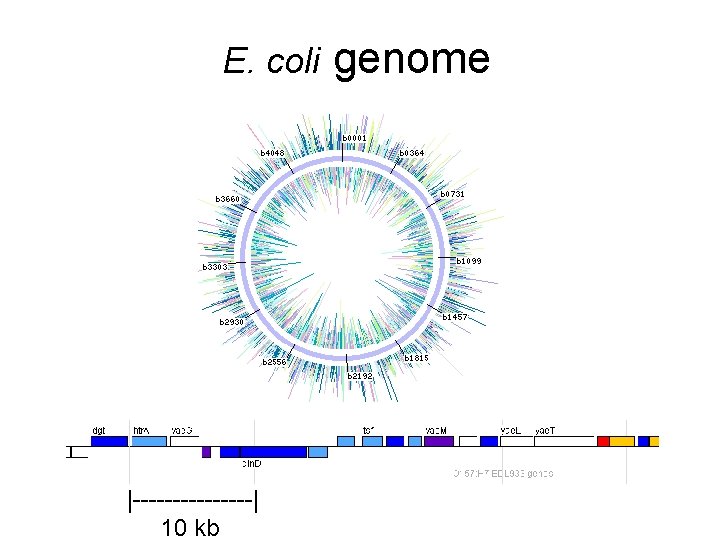
E. coli |--------| 10 kb genome
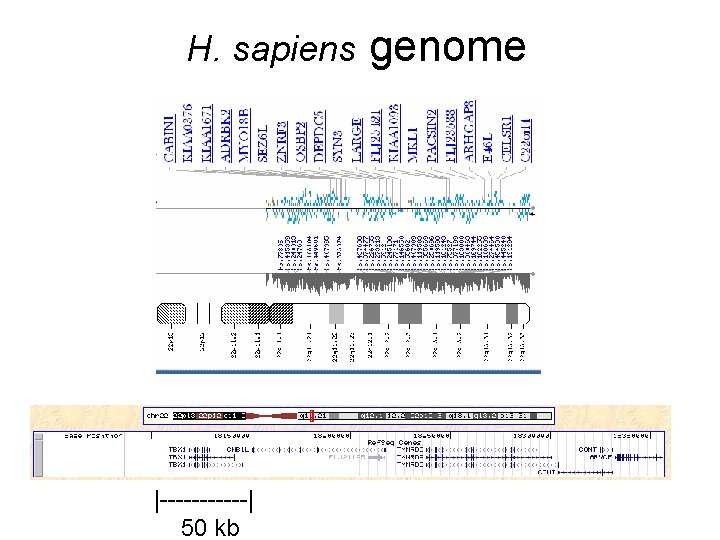
H. sapiens |------| 50 kb genome

Genome Size and Gene Number in Prokaryotes ~LINEAR

Genome Size and Gene Number in Eukaryotes Mammals
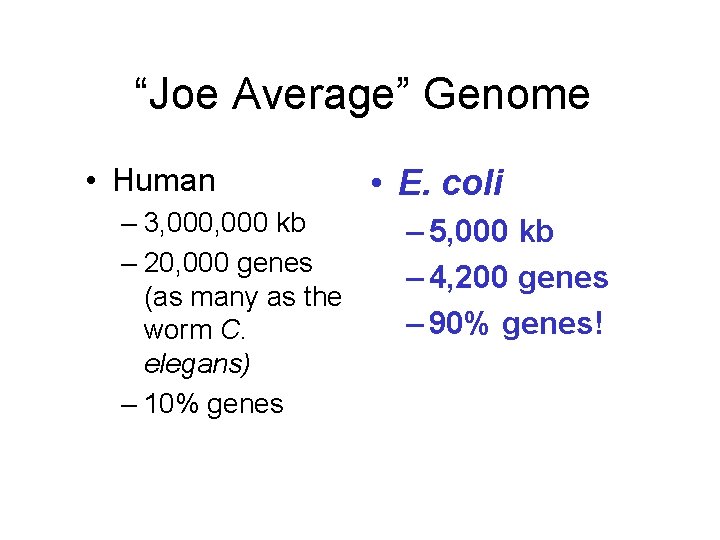
“Joe Average” Genome • Human – 3, 000 kb – 20, 000 genes (as many as the worm C. elegans) – 10% genes • E. coli – 5, 000 kb – 4, 200 genes – 90% genes!

Commonly used molecular biology techniques
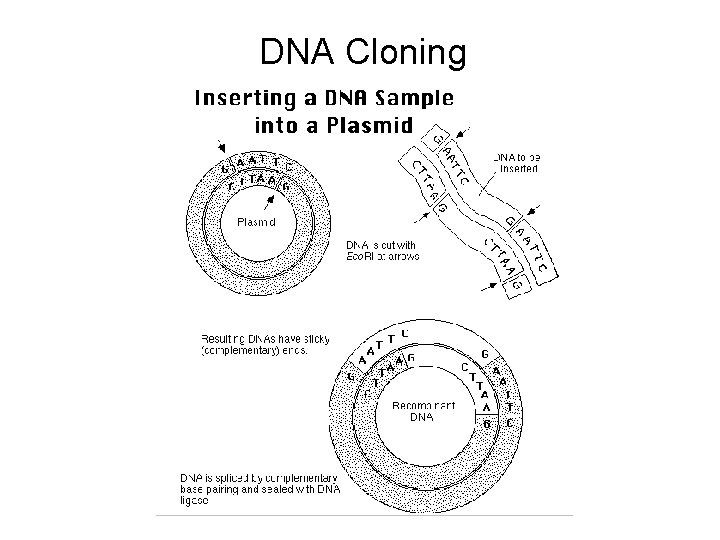
DNA Cloning

The cloning process • • Source of Nucleic Acid Cloning Vector Host Selection/Screen
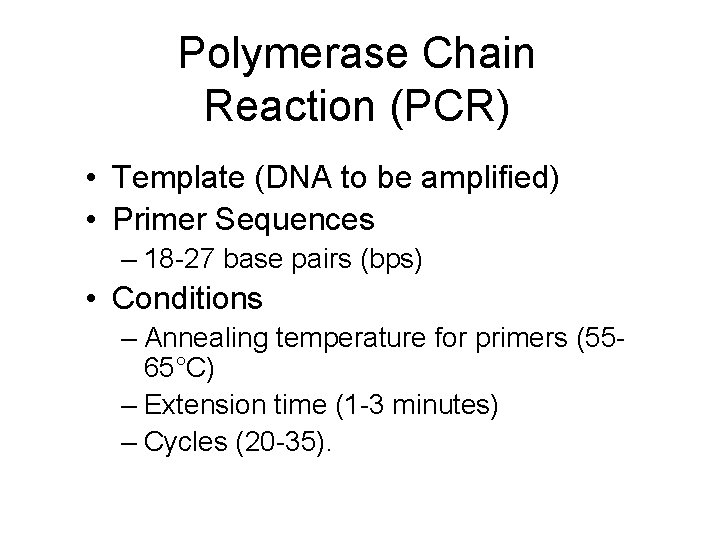
Polymerase Chain Reaction (PCR) • Template (DNA to be amplified) • Primer Sequences – 18 -27 base pairs (bps) • Conditions – Annealing temperature for primers (5565°C) – Extension time (1 -3 minutes) – Cycles (20 -35).
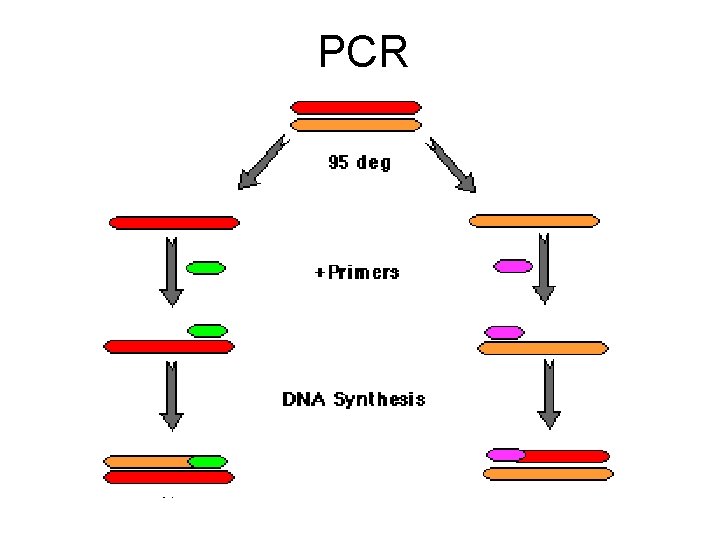
PCR
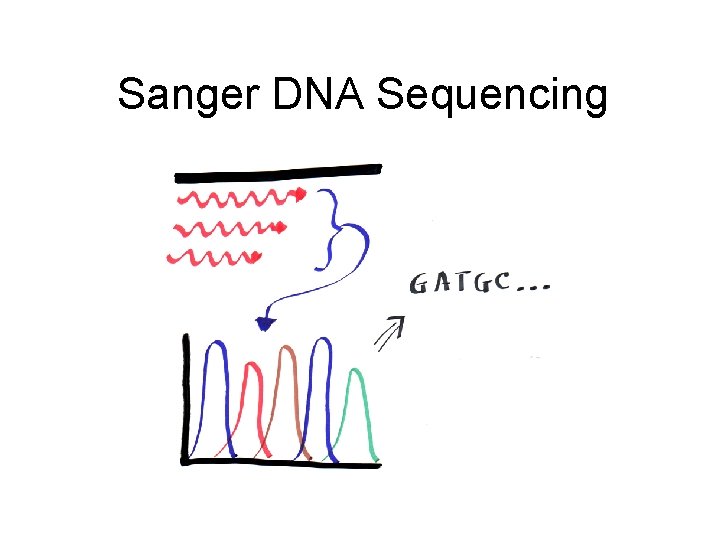
Sanger DNA Sequencing
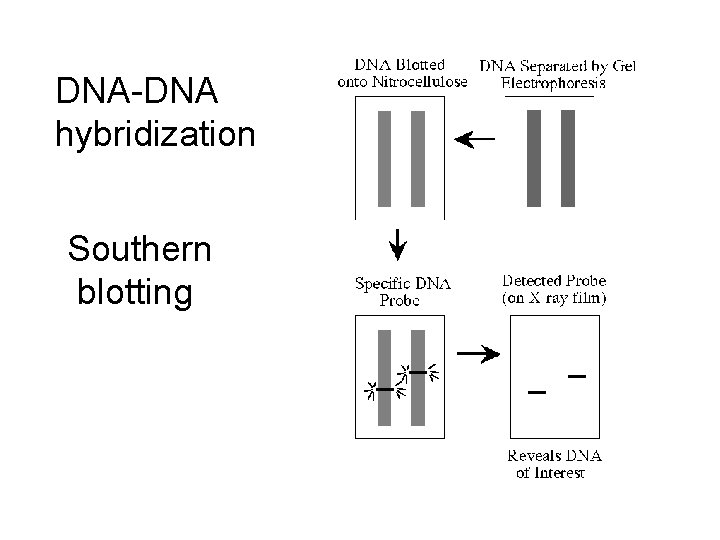
DNA-DNA hybridization Southern blotting
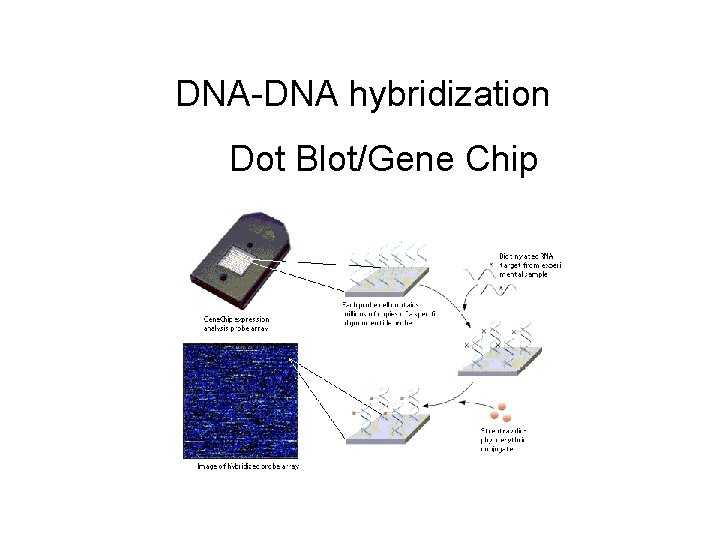
DNA-DNA hybridization Dot Blot/Gene Chip
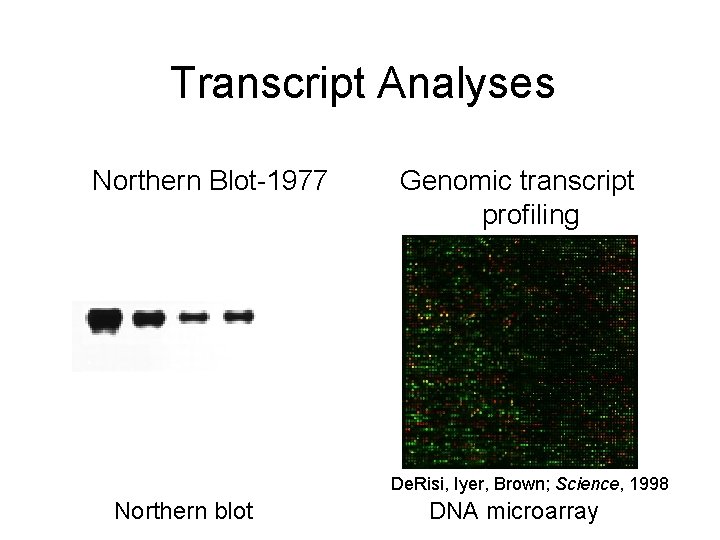
Transcript Analyses Northern Blot-1977 Genomic transcript profiling De. Risi, Iyer, Brown; Science, 1998 Northern blot DNA microarray
Questions addressed by protein analysis techniques • Identity/Amount – modifications • Structure – 3 D – Detailed information about binding to ligands and other proteins • Location • Function – Protein-protein interaction – Enzymatic activity
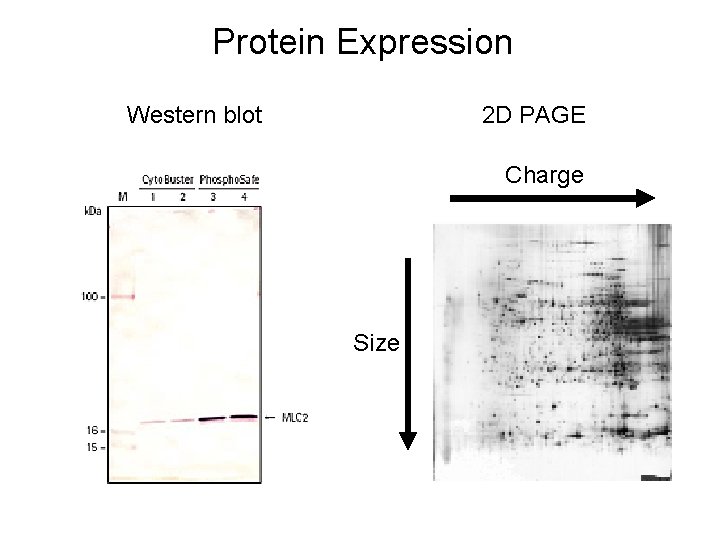
Protein Expression Western blot 2 D PAGE Charge Size
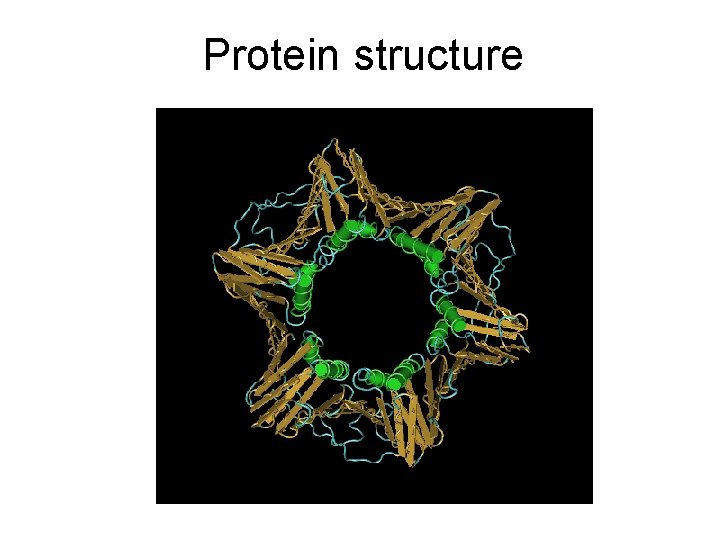
Protein structure
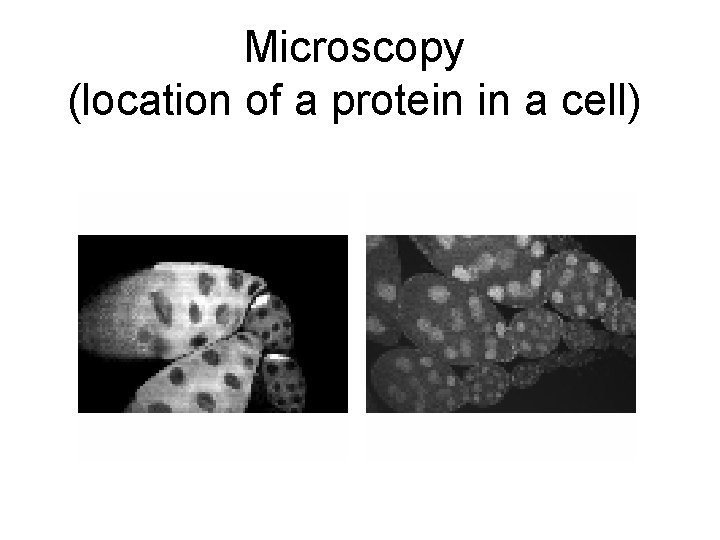
Microscopy (location of a protein in a cell)
Investigating gene function • Biochemistry • Variation – Sequence – Expression • Where, when, expression level? • Knockout • Overexpression • Homology to well-studied proteins.
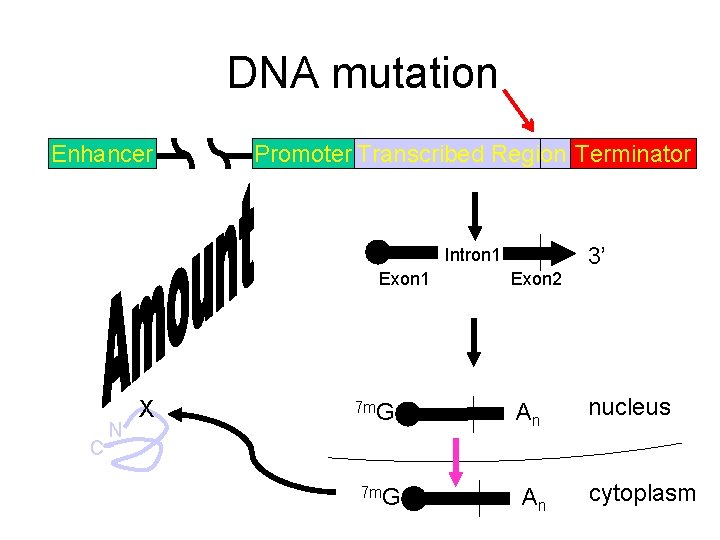
DNA mutation Enhancer Promoter Transcribed Region Terminator Intron 1 Exon 1 C N X 7 m. G Exon 2 3’ An nucleus An cytoplasm
Variation-Population • Most variation unimportant to function – SNPs (Single Nucleotide Polymorphism) • Some variation has functional importance – Eg. HBA 1 mutants • Thalassemia • Sickle Cell Anemia
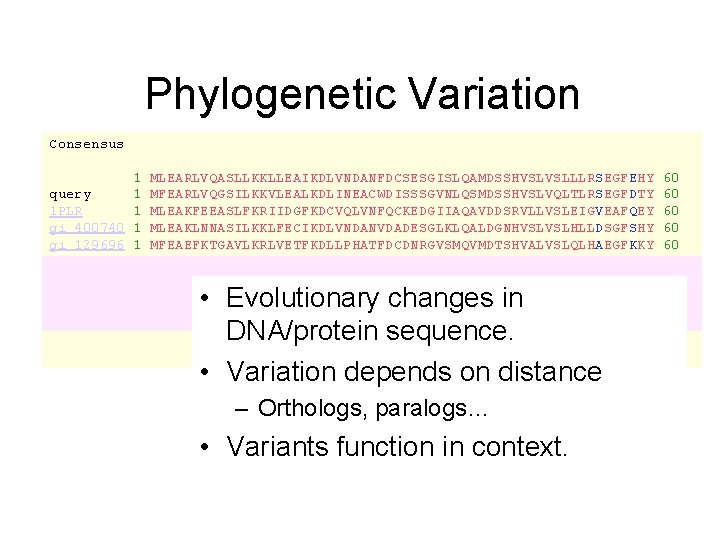
Phylogenetic Variation Consensus 1 query 1 1 PLR 1 gi 400740 1 gi 129696 1 MLEARLVQASLLKKLLEAIKDLVNDANFDCSESGISLQAMDSSHVSLVSLLLRSEGFEHY MFEARLVQGSILKKVLEALKDLINEACWDISSSGVNLQSMDSSHVSLVQLTLRSEGFDTY MLEAKFEEASLFKRIIDGFKDCVQLVNFQCKEDGIIAQAVDDSRVLLVSLEIGVEAFQEY MLEAKLNNASILKKLFECIKDLVNDANVDADESGLKLQALDGNHVSLVSLHLLDSGFSHY MFEAEFKTGAVLKRLVETFKDLLPHATFDCDNRGVSMQVMDTSHVALVSLQLHAEGFKKY • Evolutionary changes in DNA/protein sequence. • Variation depends on distance – Orthologs, paralogs… • Variants function in context. 60 60 60
Induced/Directed Mutation “Experimental” • Often GROSS mutations • May not reflect in any way the population or phylogenetic variation • Critical to functional study
- Slides: 47