Biological Functions of Glycosylation Cell Recognition Protein Folding


Biological Functions of Glycosylation Cell Recognition Protein Folding Glycosylation 糖 链 糖链 Immunity Cell Adhesion Reproduction
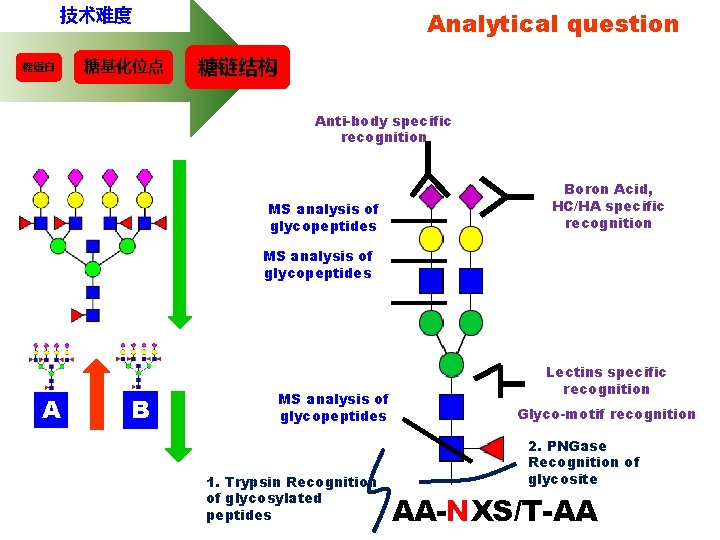
技术难度 糖蛋白 糖基化位点 Analytical question 糖链结构 Anti-body specific recognition MS analysis of glycopeptides Boron Acid, HC/HA specific recognition MS analysis of glycopeptides A B MS analysis of glycopeptides 1. Trypsin Recognition of glycosylated peptides Lectins specific recognition Glyco-motif recognition 2. PNGase Recognition of glycosite AA-NXS/T-AA
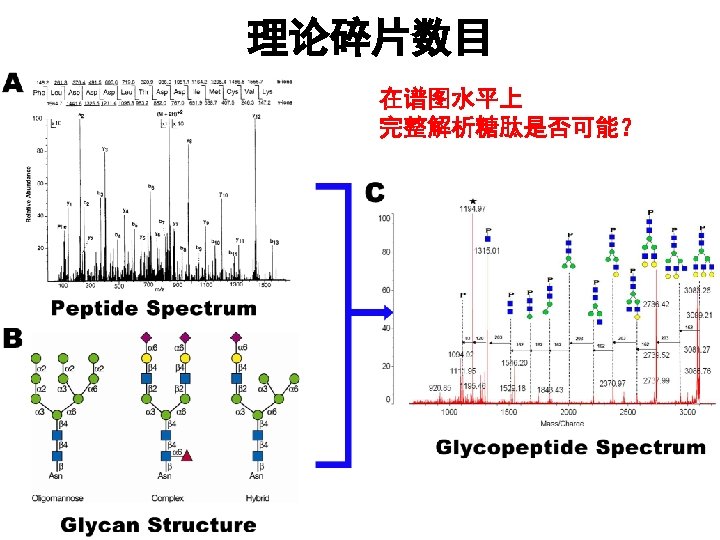
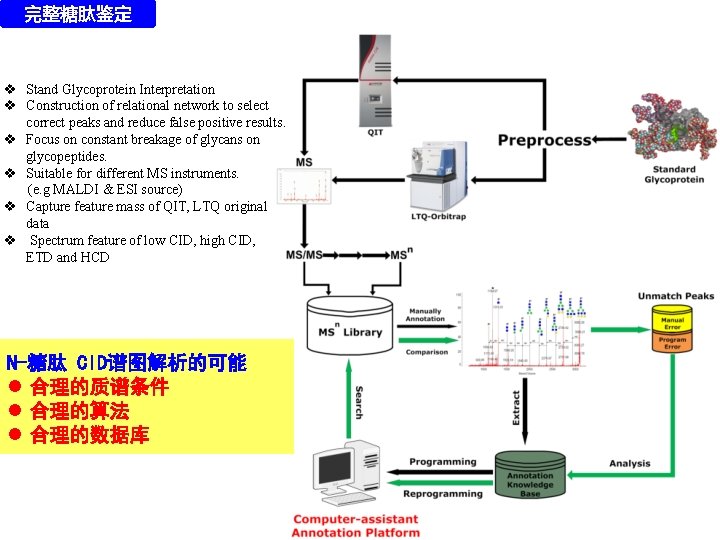
完整糖肽鉴定 v Stand Glycoprotein Interpretation v Construction of relational network to select correct peaks and reduce false positive results. v Focus on constant breakage of glycans on glycopeptides. v Suitable for different MS instruments. (e. g MALDI & ESI source) v Capture feature mass of QIT, LTQ original data v Spectrum feature of low CID, high CID, ETD and HCD N-糖肽 CID谱图解析的可能 l 合理的质谱条件 l 合理的算法 l 合理的数据库
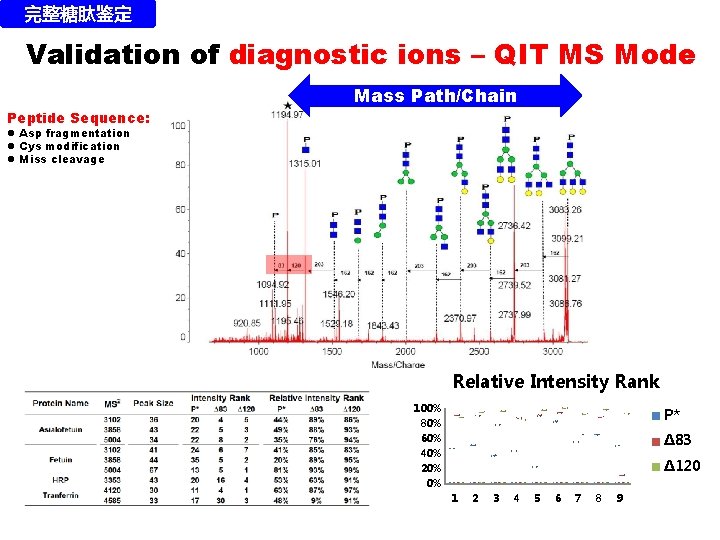
完整糖肽鉴定 Validation of diagnostic ions – QIT MS Mode Mass Path/Chain Peptide Sequence: l Asp fragmentation l Cys modification l Miss cleavage Relative Intensity Rank 100% 80% 60% 40% 20% 0% P* ∆83 ∆120 1 2 3 4 5 6 7 8 9
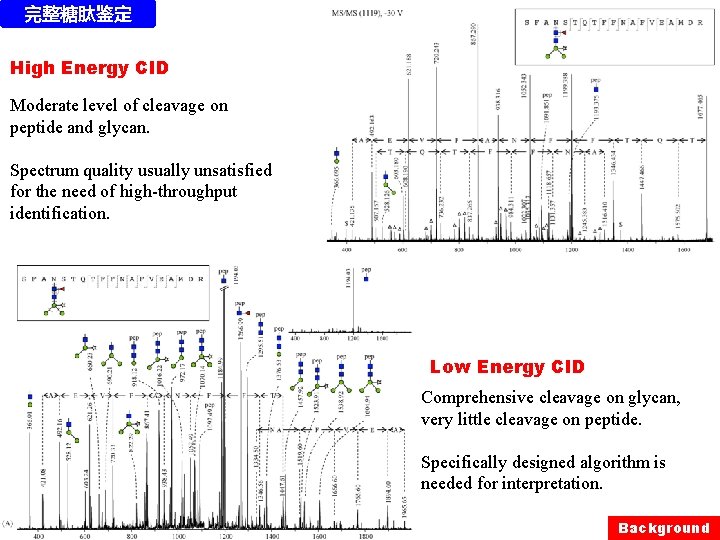
完整糖肽鉴定 High Energy CID Moderate level of cleavage on peptide and glycan. Spectrum quality usually unsatisfied for the need of high-throughput identification. Low Energy CID Comprehensive cleavage on glycan, very little cleavage on peptide. Specifically designed algorithm is needed for interpretation. Background
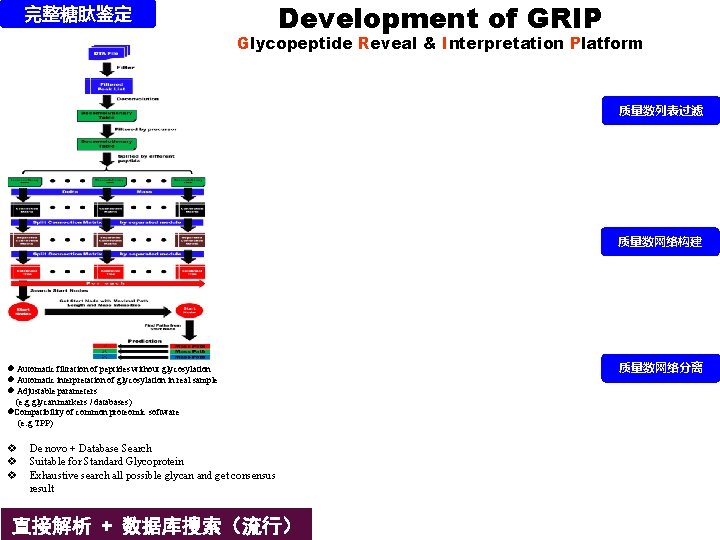
完整糖肽鉴定 Development of GRIP Glycopeptide Reveal & Interpretation Platform 质量数列表过滤 质量数网络构建 l Automatic filtration of peptides without glycosylation l Automatic interpretation of glycosylation in real sample l Adjustable parameters (e. g glycan markers / databases) l. Compatibility of common proteomic software (e. g TPP) v v v De novo + Database Search Suitable for Standard Glycoprotein Exhaustive search all possible glycan and get consensus result 直接解析 + 数据库搜索(流行) 质量数网络分离
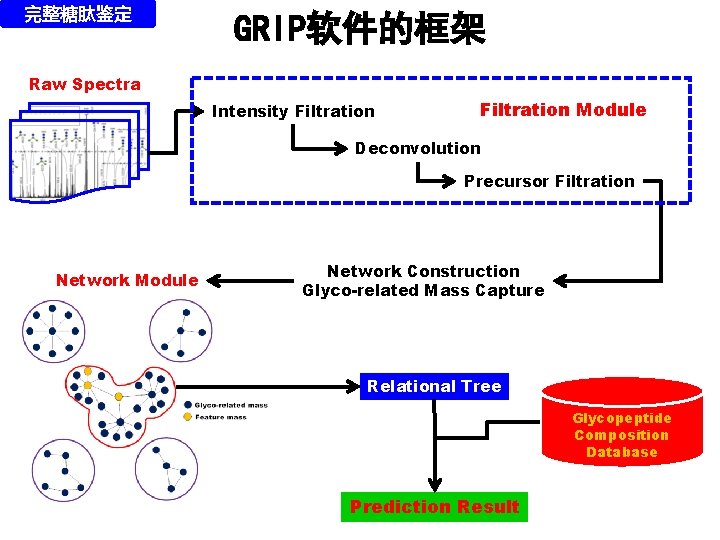
完整糖肽鉴定 GRIP软件的框架 Raw Spectra Intensity Filtration Module Deconvolution Precursor Filtration Network Module Network Construction Glyco-related Mass Capture Relational Tree Glycopeptide Composition Database Prediction Result
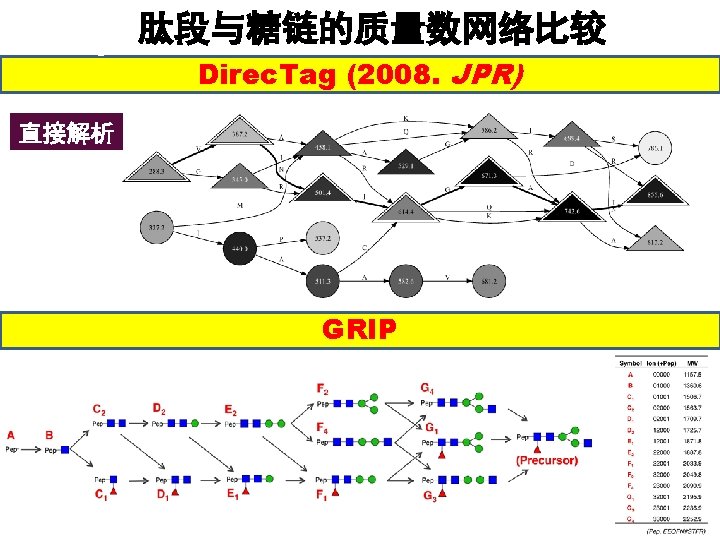
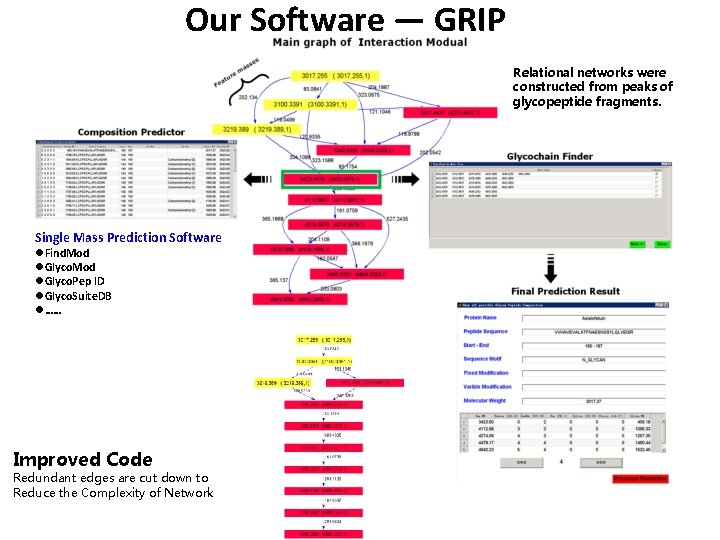
Our Software — GRIP Relational networks were constructed from peaks of glycopeptide fragments. Single Mass Prediction Software l. Find. Mod l. Glyco. Pep ID l. Glyco. Suite. DB l…… Improved Code Redundant edges are cut down to Reduce the Complexity of Network
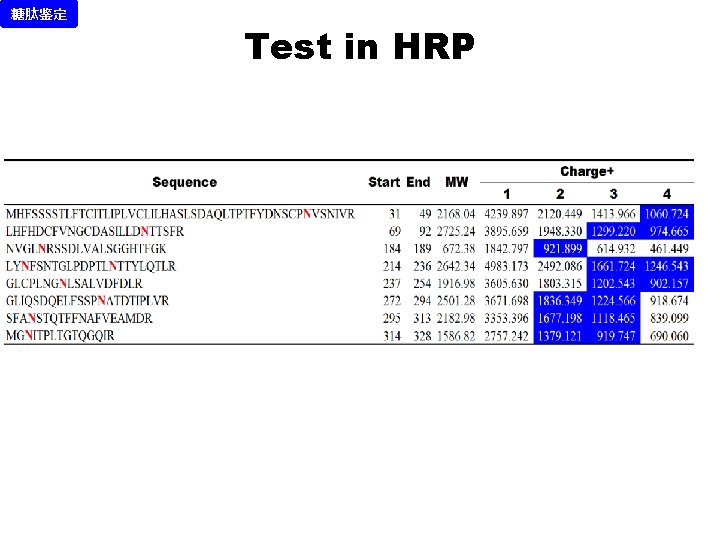
糖肽鉴定 Test in HRP
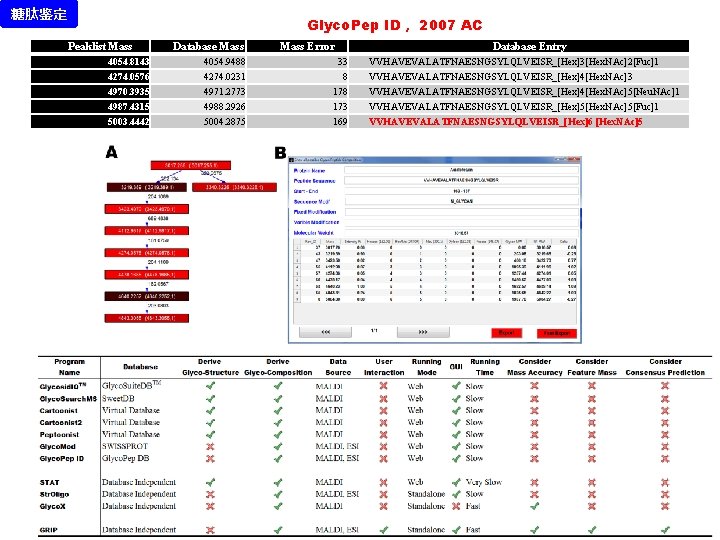
糖肽鉴定 Glyco. Pep ID, 2007 AC Peaklist Mass 4054. 8143 4274. 0576 4970. 3935 4987. 4315 5003. 4442 Database Mass 4054. 9488 4274. 0231 4971. 2773 4988. 2926 5004. 2875 Mass Error 33 8 173 169 Database Entry VVHAVEVALATFNAESNGSYLQLVEISR_[Hex]3[Hex. NAc]2[Fuc]1 VVHAVEVALATFNAESNGSYLQLVEISR_[Hex]4[Hex. NAc]3 VVHAVEVALATFNAESNGSYLQLVEISR_[Hex]4[Hex. NAc]5[Neu. NAc]1 VVHAVEVALATFNAESNGSYLQLVEISR_[Hex]5[Hex. NAc]5[Fuc]1 VVHAVEVALATFNAESNGSYLQLVEISR_[Hex]6 [Hex. NAc]5
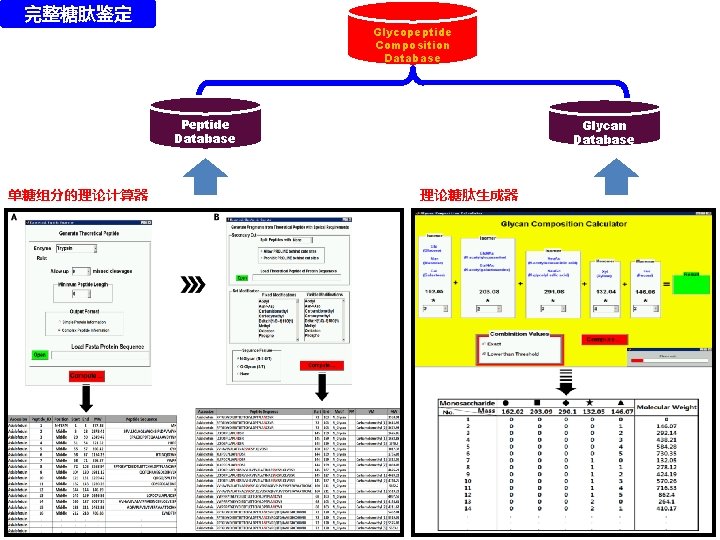
完整糖肽鉴定 Glycopeptide Composition Database Peptide Database 单糖组分的理论计算器 Glycan Database 理论糖肽生成器
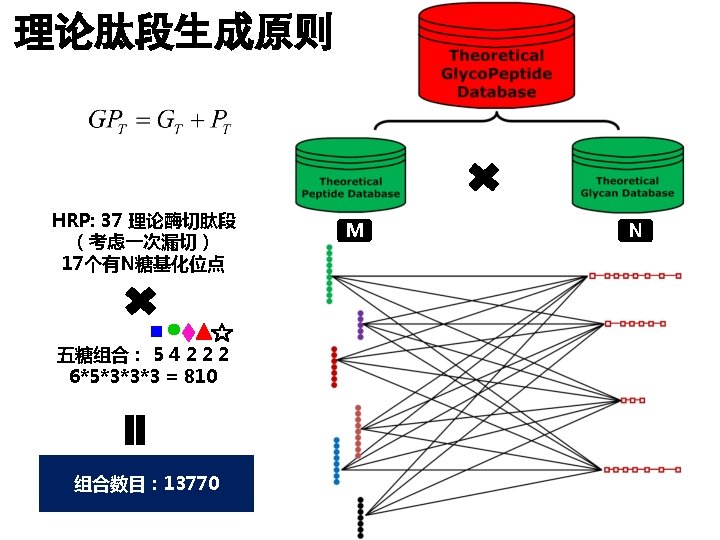
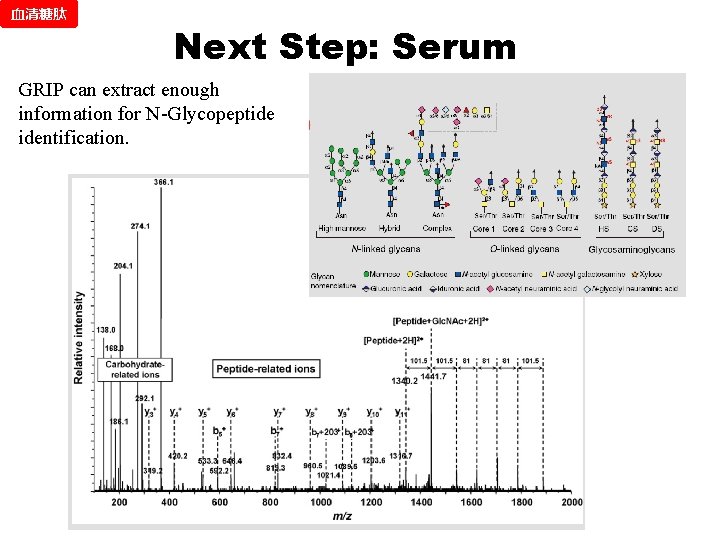
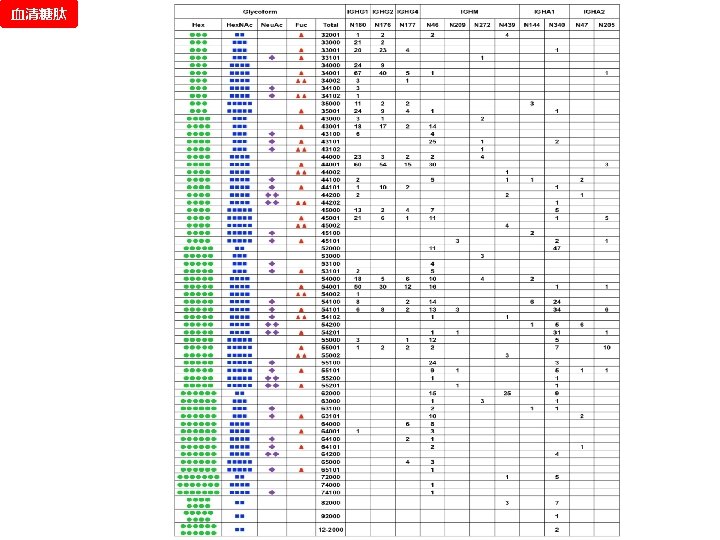
血清糖肽 Next Step: Serum GRIP can extract enough information for N-Glycopeptide identification.
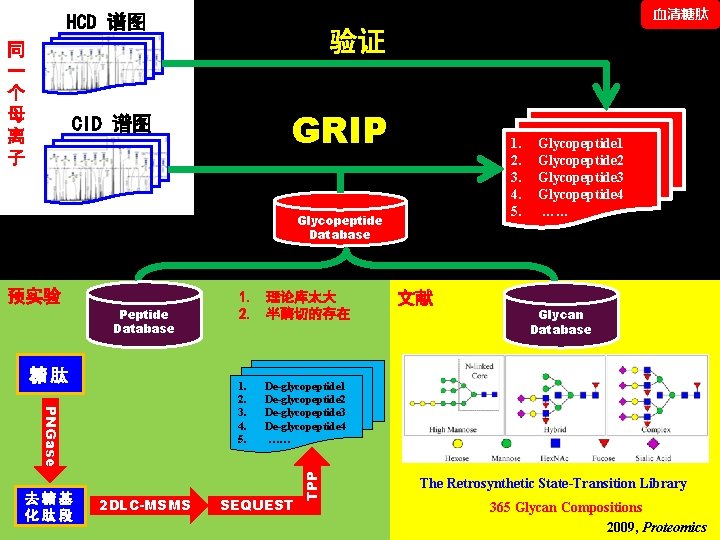
血清糖肽 HCD 谱图 同 一 个 母 离 子 验证 GRIP CID 谱图 1. 2. 3. 4. 5. Glycopeptide Database Peptide Database 糖肽 1. 2. 3. 4. 5. PNGase 去糖基 化肽段 1. 理论库太大 2. 半酶切的存在 2 DLC-MSMS 文献 Glycan Database De-glycopeptide 1 De-glycopeptide 2 De-glycopeptide 3 De-glycopeptide 4 …… SEQUEST TPP 预实验 Glycopeptide 1 Glycopeptide 2 Glycopeptide 3 Glycopeptide 4 …… The Retrosynthetic State-Transition Library 365 Glycan Compositions 2009, Proteomics
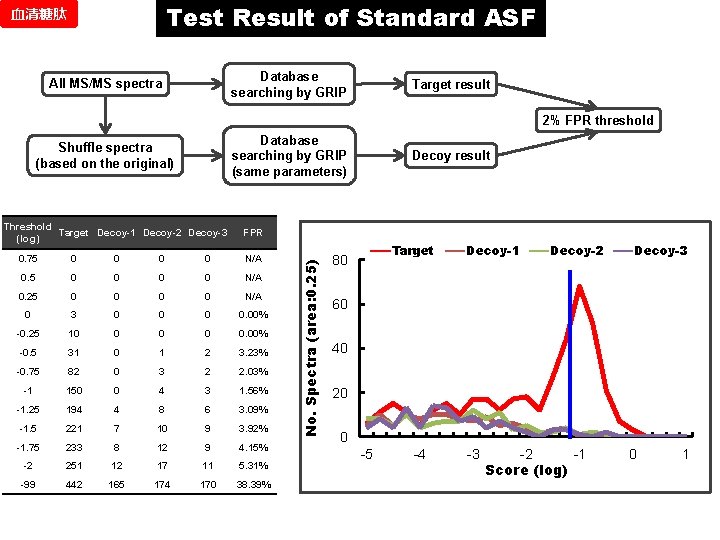
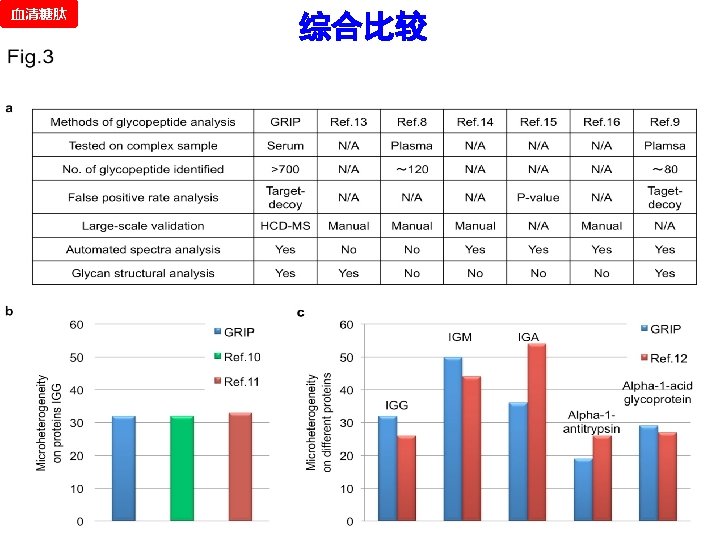
Test Result of Standard ASF 血清糖肽 Database searching by GRIP All MS/MS spectra Target result 2% FPR threshold Database searching by GRIP (same parameters) Threshold Target Decoy-1 Decoy-2 Decoy-3 (log) Decoy result FPR 0. 75 0 0 0 0 N/A 0. 25 0 0 N/A 0 3 0 0. 00% -0. 25 10 0 0. 00% -0. 5 31 0 1 2 3. 23% -0. 75 82 0 3 2 2. 03% -1 150 0 4 3 1. 56% -1. 25 194 4 8 6 3. 09% -1. 5 221 7 10 9 3. 92% -1. 75 233 8 12 9 4. 15% -2 251 12 17 11 5. 31% -99 442 165 174 170 38. 39% No. Spectra (area: 0. 25) Shuffle spectra (based on the original) Target 80 Decoy-1 Decoy-2 Decoy-3 60 40 20 0 -5 -4 -3 -2 -1 Score (log) 0 1
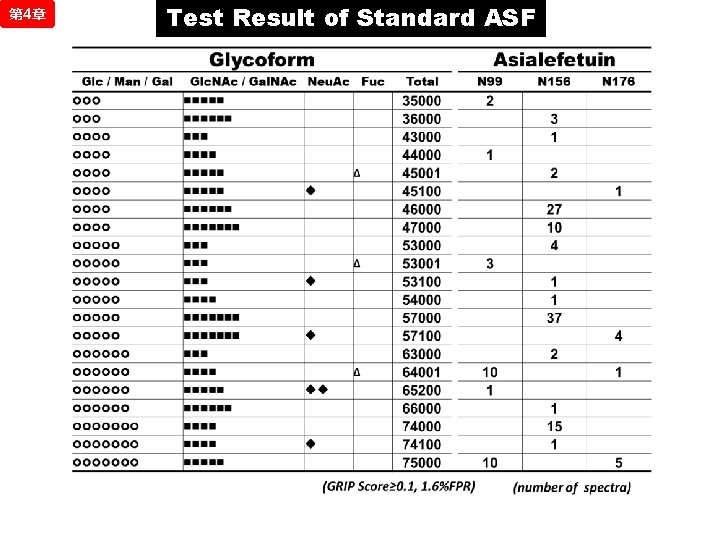
第 4章 Test Result of Standard ASF
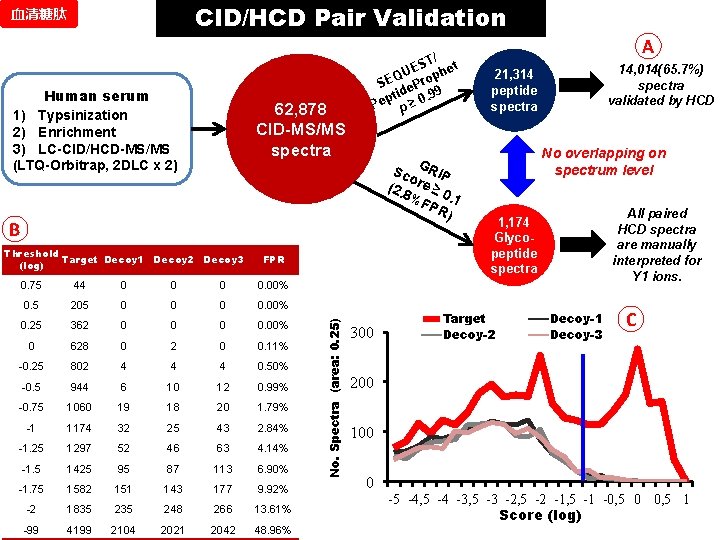
CID/HCD Pair Validation 血清糖肽 Human serum 1) Typsinization 2) Enrichment 3) LC-CID/HCD-MS/MS (LTQ-Orbitrap, 2 DLC x 2) 62, 878 CID-MS/MS spectra / ST et E ph QU SE e. Pro d. 99 pti Pe p ≥ 0 Sc GRIP o (2. 8 re ≥ %F 0. 1 PR ) B FPR 0. 75 44 0 0. 00% 0. 5 205 0 0. 00% 0. 25 362 0 0. 00% 0 628 0 2 0 0. 11% -0. 25 802 4 4 4 0. 50% -0. 5 944 6 10 12 0. 99% -0. 75 1060 19 18 20 1. 79% -1 1174 32 25 43 2. 84% -1. 25 1297 52 46 63 4. 14% -1. 5 1425 95 87 113 6. 90% -1. 75 1582 151 143 177 9. 92% -2 1835 248 266 13. 61% -99 4199 2104 2021 2042 48. 96% No. Spectra (area: 0. 25) Threshold Target Decoy 1 Decoy 2 Decoy 3 (log) 300 A 14, 014(65. 7%) spectra validated by HCD 21, 314 peptide spectra No overlapping on spectrum level All paired HCD spectra are manually interpreted for Y 1 ions. 1, 174 Glycopeptide spectra Target Decoy-2 Decoy-1 Decoy-3 C 200 100 0 -5 -4, 5 -4 -3, 5 -3 -2, 5 -2 -1, 5 -1 -0, 5 0 0, 5 1 Score (log)
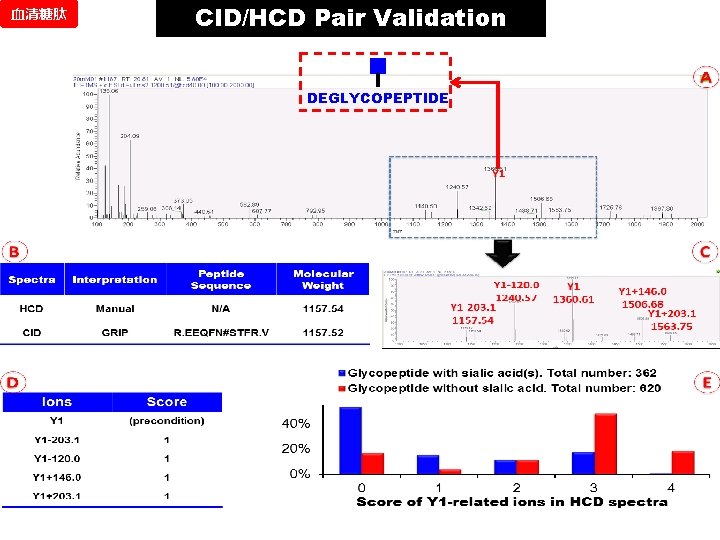
血清糖肽 CID/HCD Pair Validation DEGLYCOPEPTIDE
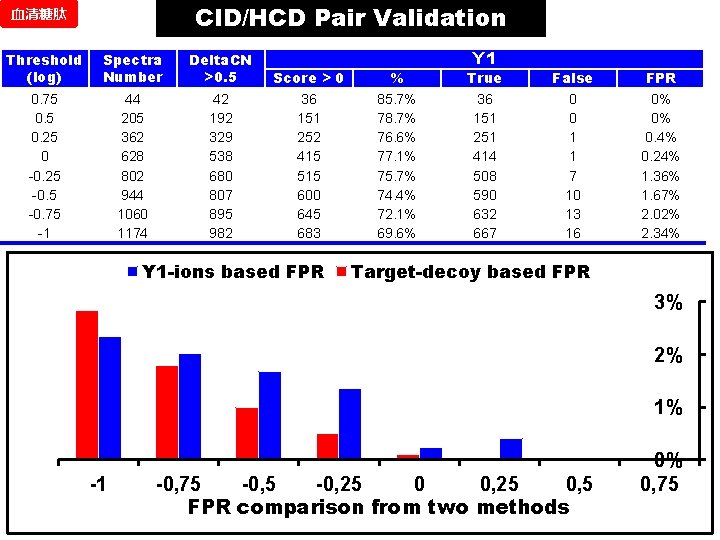
CID/HCD Pair Validation 血清糖肽 Threshold (log) Spectra Number Delta. CN >0. 5 0. 75 0. 25 0 -0. 25 -0. 75 -1 44 205 362 628 802 944 1060 1174 42 192 329 538 680 807 895 982 Score > 0 36 151 252 415 515 600 645 683 Y 1 -ions based FPR % 85. 7% 78. 7% 76. 6% 77. 1% 75. 7% 74. 4% 72. 1% 69. 6% Y 1 True 36 151 251 414 508 590 632 667 False 0 0 1 1 7 10 13 16 FPR 0% 0% 0. 4% 0. 24% 1. 36% 1. 67% 2. 02% 2. 34% Target-decoy based FPR 3% 2% 1% -1 -0, 75 -0, 25 0, 5 FPR comparison from two methods 0% 0, 75
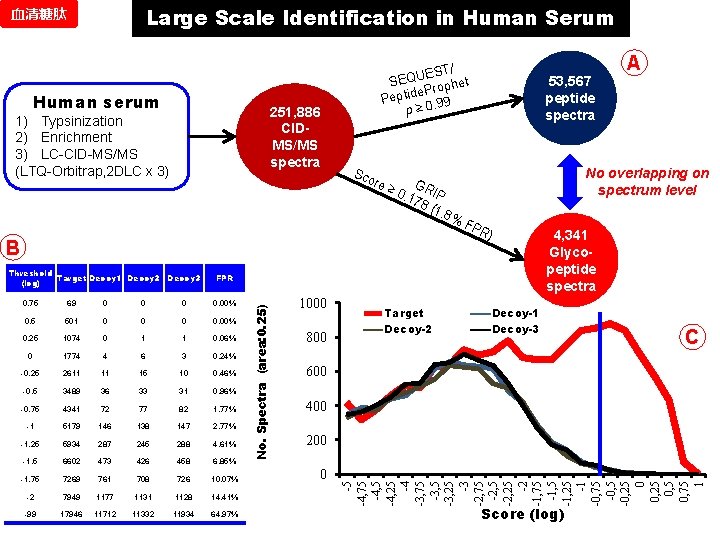
Large Scale Identification in Human Serum 血清糖肽 Human serum 251, 886 CIDMS/MS spectra 1) Typsinization 2) Enrichment 3) LC-CID-MS/MS (LTQ-Orbitrap, 2 DLC x 3) EST/ SEQU ophet e. Pr Peptid 0. 99 p≥ Sco re ≥ G 0. 1 RIP 78 (1. 8 % Decoy 3 FPR 0. 75 69 0 0. 00% 0. 5 501 0 0. 00% 0. 25 1074 0 1 1 0. 06% 0 1774 4 6 3 0. 24% -0. 25 2611 11 15 10 0. 46% -0. 5 3489 36 33 31 0. 96% -0. 75 4341 72 77 82 1. 77% -1 5179 146 138 147 2. 77% -1. 25 5934 287 245 288 4. 61% -1. 5 6602 473 426 458 6. 85% -1. 75 7269 761 708 726 10. 07% -2 7949 1177 1131 1128 14. 41% -99 17946 11712 11332 11934 64. 97% 1000 800 Target Decoy-2 No overlapping on spectrum level FP R) 4, 341 Glycopeptide spectra Decoy-1 Decoy-3 C 600 400 200 0 -5 -4, 75 -4, 25 -4 -3, 75 -3, 25 -3 -2, 75 -2, 25 -2 -1, 75 -1, 25 -1 -0, 75 -0, 25 0, 75 1 Threshold Target Decoy 1 Decoy 2 (log) No. Spectra (area: 0. 25) B 53, 567 peptide spectra A Score (log)
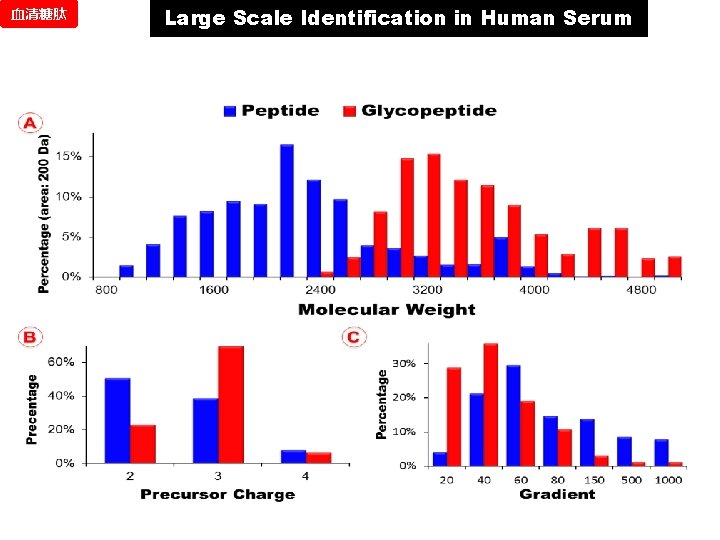
血清糖肽 Large Scale Identification in Human Serum
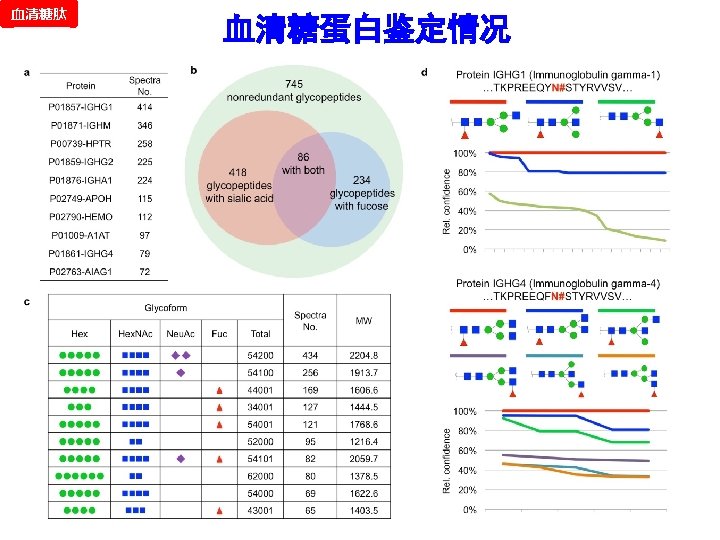
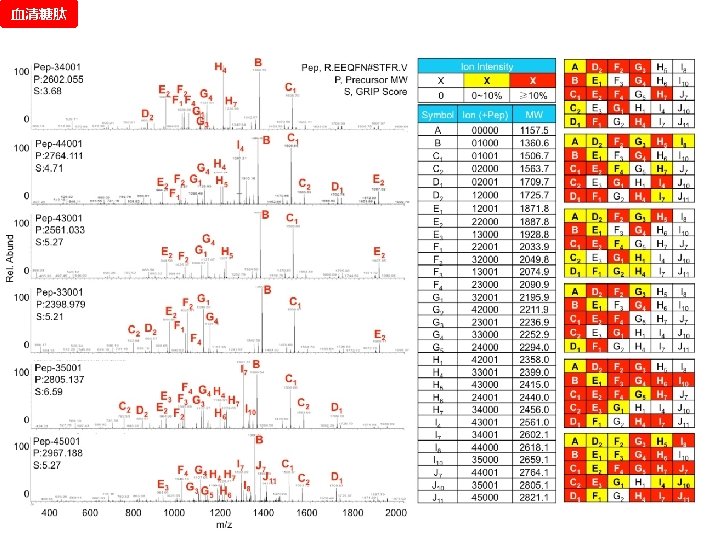
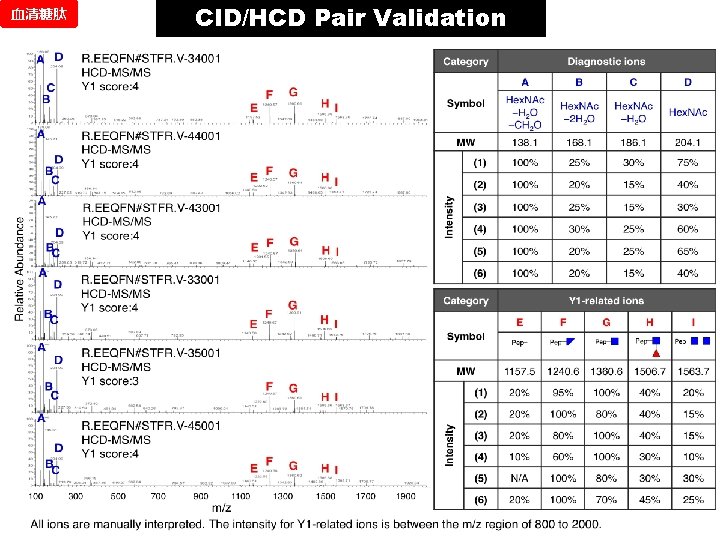
血清糖肽 CID/HCD Pair Validation
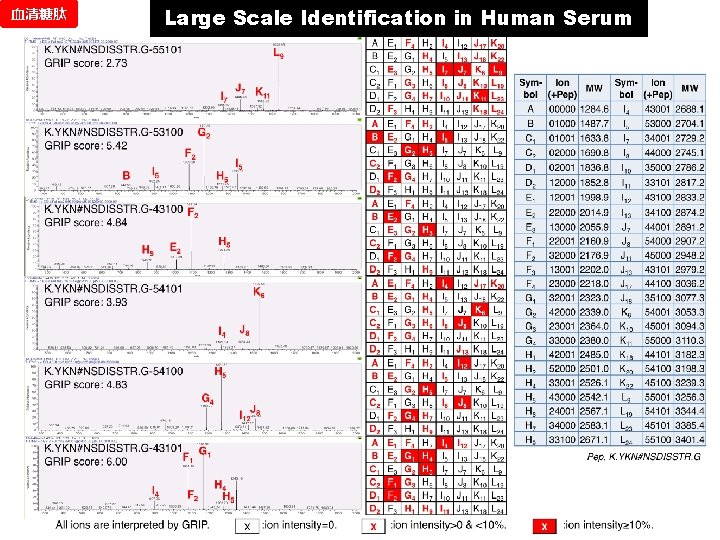
血清糖肽 Large Scale Identification in Human Serum
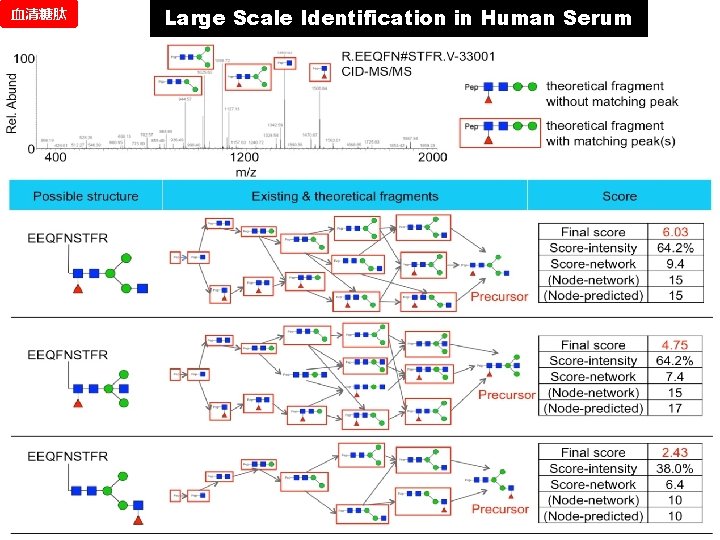
血清糖肽 Large Scale Identification in Human Serum
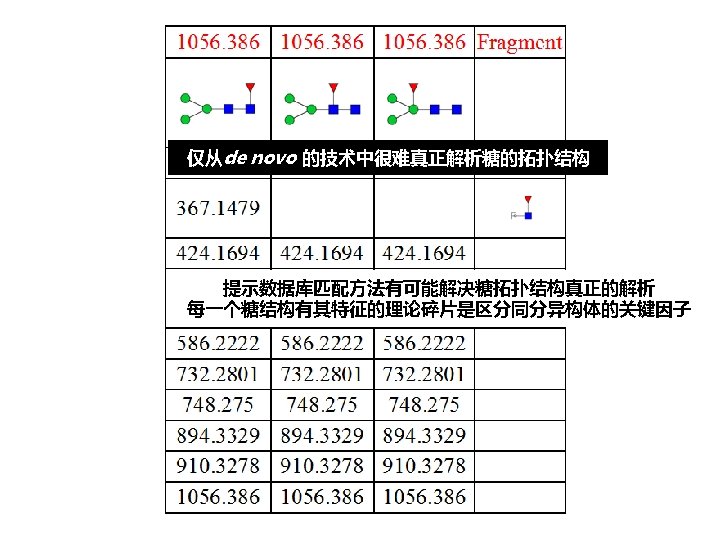
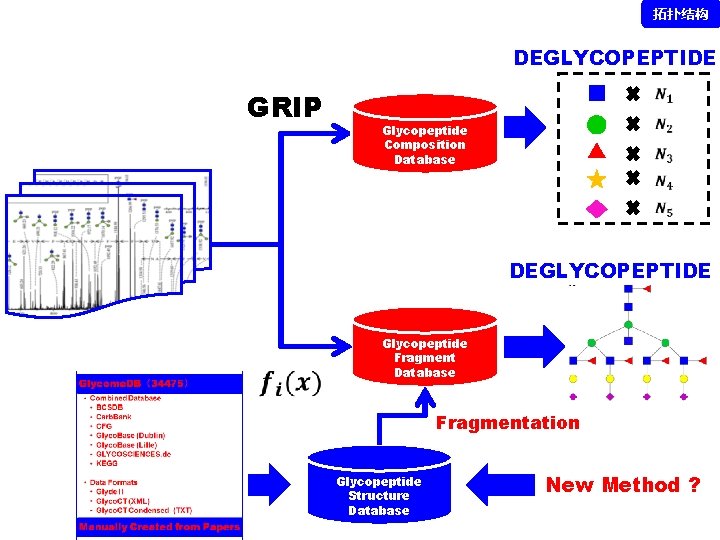
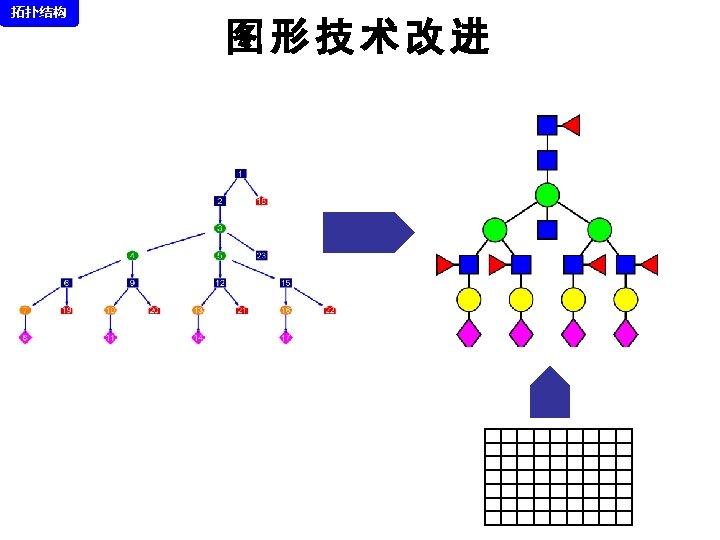
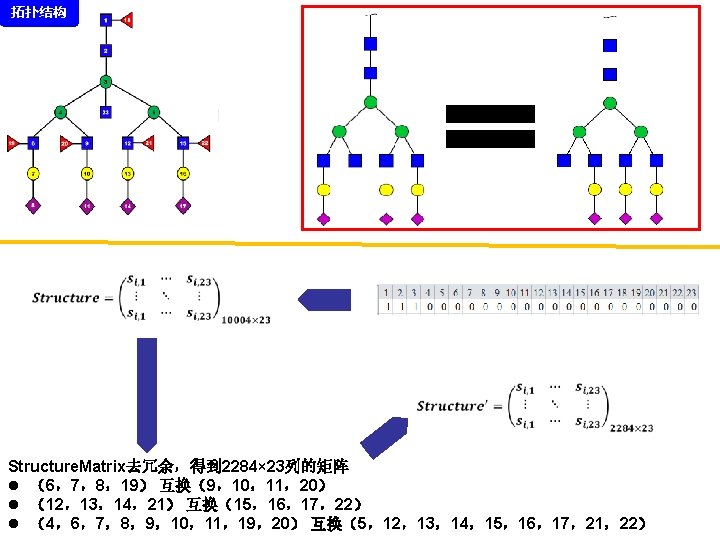
拓扑结构 DEGLYCOPEPTIDE GRIP Glycopeptide Composition Database DEGLYCOPEPTIDE Glycopeptide Fragment Database Fragmentation Glycopeptide Structure Database New Method ?
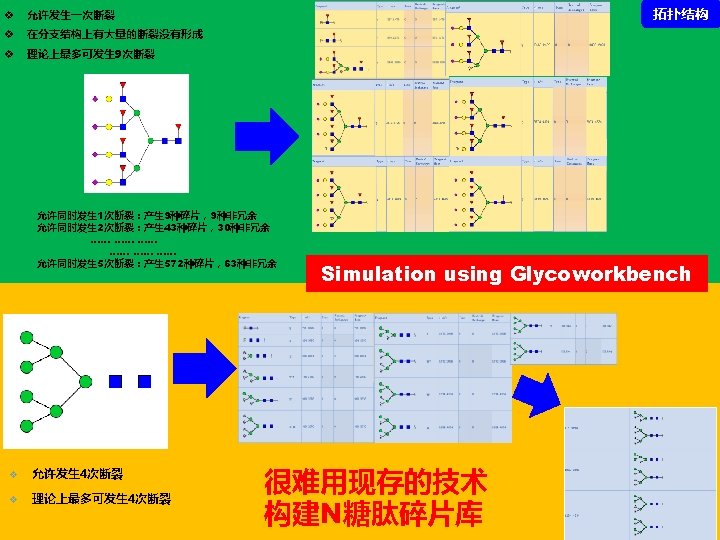
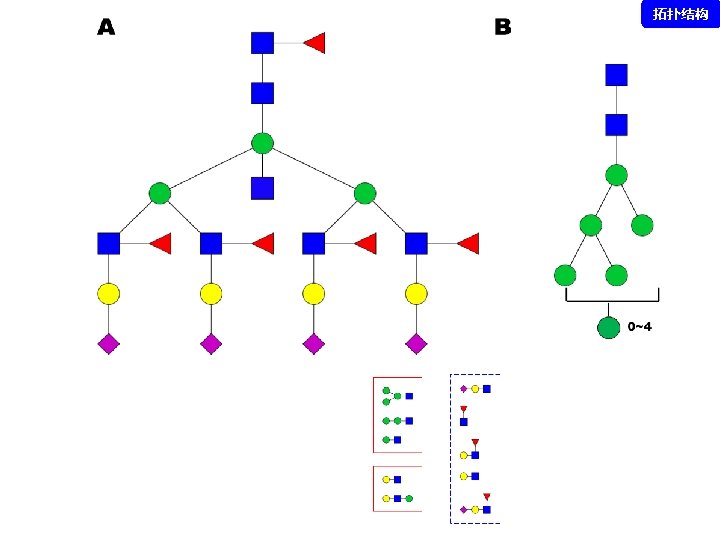
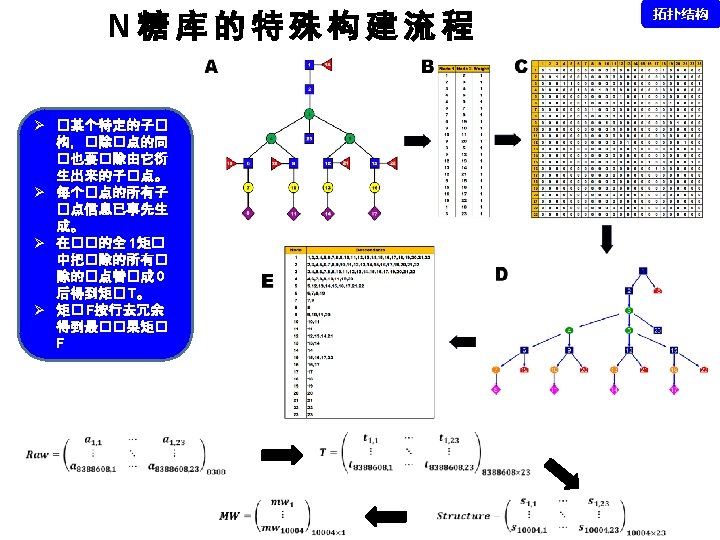
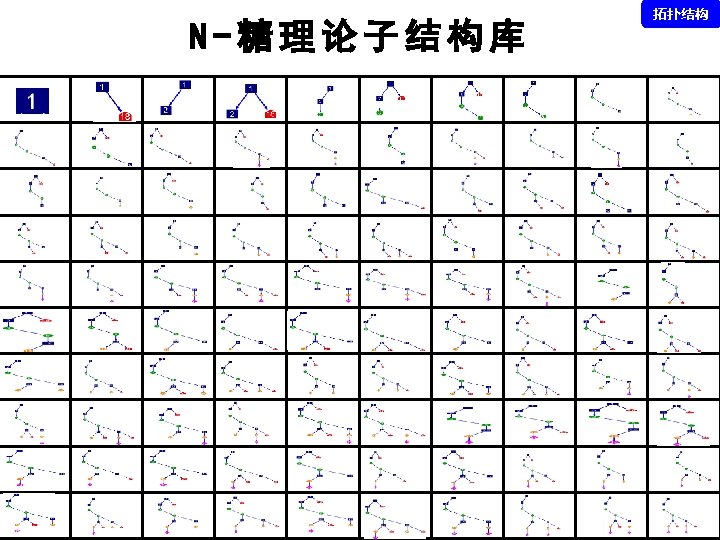
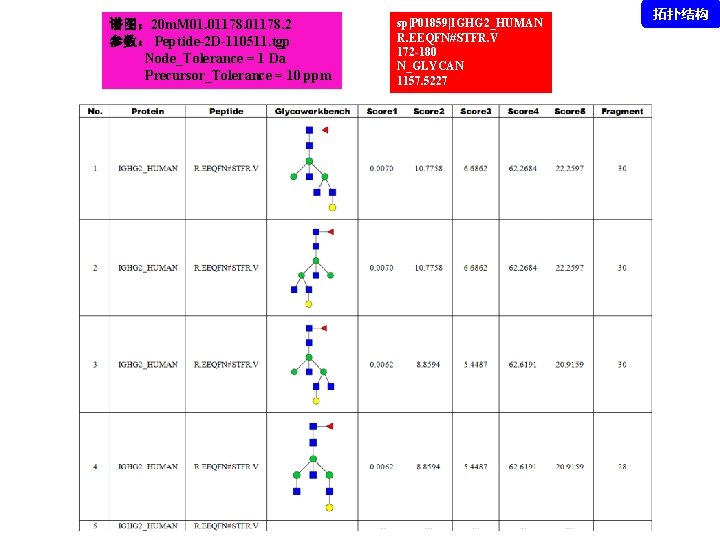
谱图: 20 m. M 01. 01178. 2 参数: Peptide-2 D-110511. tgp Node_Tolerance = 1 Da Precursor_Tolerance = 10 ppm sp|P 01859|IGHG 2_HUMAN R. EEQFN#STFR. V 172 -180 N_GLYCAN 1157. 5227 拓扑结构


- Slides: 46