BioLinux An integrated bioinformatics solution for the EG
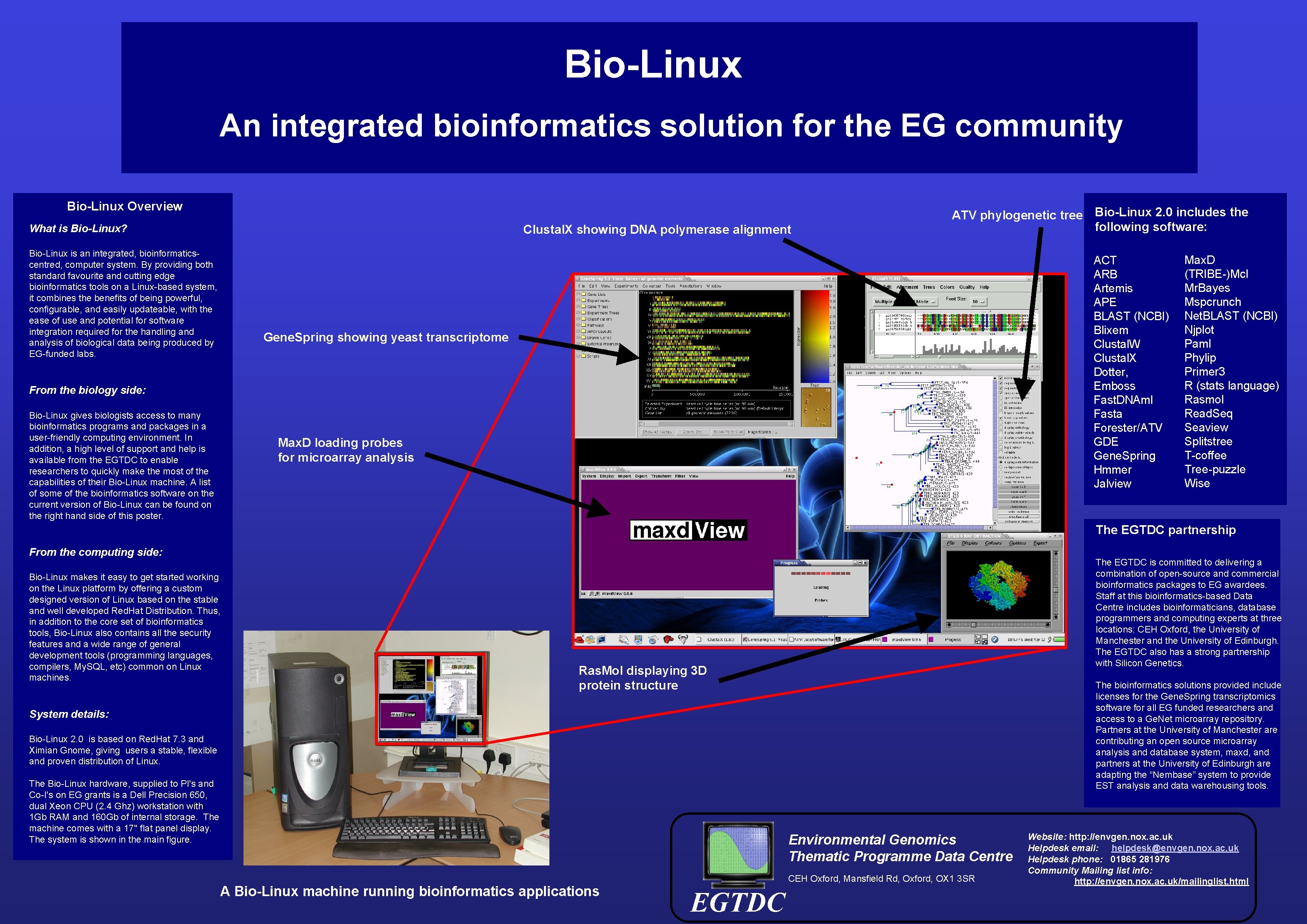
Bio-Linux An integrated bioinformatics solution for the EG community Bio-Linux Overview What is Bio-Linux? Bio-Linux is an integrated, bioinformaticscentred, computer system. By providing both standard favourite and cutting edge bioinformatics tools on a Linux-based system, it combines the benefits of being powerful, configurable, and easily updateable, with the ease of use and potential for software integration required for the handling and analysis of biological data being produced by EG-funded labs. Clustal. X showing DNA polymerase alignment ATV phylogenetic tree Bio-Linux 2. 0 includes the following software: ACT ARB Artemis APE BLAST (NCBI) Blixem Clustal. W Clustal. X Dotter, Emboss Fast. DNAml Fasta Forester/ATV GDE Gene. Spring Hmmer Jalview Gene. Spring showing yeast transcriptome From the biology side: Bio-Linux gives biologists access to many bioinformatics programs and packages in a user-friendly computing environment. In addition, a high level of support and help is available from the EGTDC to enable researchers to quickly make the most of the capabilities of their Bio-Linux machine. A list of some of the bioinformatics software on the current version of Bio-Linux can be found on the right hand side of this poster. Max. D loading probes for microarray analysis Max. D (TRIBE-)Mcl Mr. Bayes Mspcrunch Net. BLAST (NCBI) Njplot Paml Phylip Primer 3 R (stats language) Rasmol Read. Seq Seaview Splitstree T-coffee Tree-puzzle Wise The EGTDC partnership From the computing side: Bio-Linux makes it easy to get started working on the Linux platform by offering a custom designed version of Linux based on the stable and well developed Red. Hat Distribution. Thus, in addition to the core set of bioinformatics tools, Bio-Linux also contains all the security features and a wide range of general development tools (programming languages, compilers, My. SQL, etc) common on Linux machines. The EGTDC is committed to delivering a combination of open-source and commercial bioinformatics packages to EG awardees. Staff at this bioinformatics-based Data Centre includes bioinformaticians, database programmers and computing experts at three locations: CEH Oxford, the University of Manchester and the University of Edinburgh. The EGTDC also has a strong partnership with Silicon Genetics. Ras. Mol displaying 3 D protein structure The bioinformatics solutions provided include licenses for the Gene. Spring transcriptomics software for all EG funded researchers and access to a Ge. Net microarray repository. Partners at the University of Manchester are contributing an open source microarray analysis and database system, maxd, and partners at the University of Edinburgh are adapting the “Nembase” system to provide EST analysis and data warehousing tools. System details: Bio-Linux 2. 0 is based on Red. Hat 7. 3 and Ximian Gnome, giving users a stable, flexible and proven distribution of Linux. The Bio-Linux hardware, supplied to PI's and Co-I's on EG grants is a Dell Precision 650, dual Xeon CPU (2. 4 Ghz) workstation with 1 Gb RAM and 160 Gb of internal storage. The machine comes with a 17'' flat panel display. The system is shown in the main figure. Environmental Genomics Thematic Programme Data Centre A Bio-Linux machine running bioinformatics applications CEH Oxford, Mansfield Rd, Oxford, OX 1 3 SR EGTDC Website: http: //envgen. nox. ac. uk Helpdesk email: helpdesk@envgen. nox. ac. uk Helpdesk phone: 01865 281976 Community Mailing list info: http: //envgen. nox. ac. uk/mailinglist. html
- Slides: 1