BioinformaticsPCR Lab How does having a certain genetic

Bioinformatics/PCR Lab How does having a certain genetic marker affect chances of getting brain cancer?

Bioinformatics/PCR Lab How does having a certain genetic marker affect chances of getting brain cancer?
Bioinformatics/PCR Lab Important Concepts: • genetic tests (molecular) • genetic marker • sequence (DNA) • RFLP (restriction fragment length polymorphism) • genetics and disease • greater risk vs. known association • population studies

Bioinformatics/PCR Lab Sampling of Genetic Tests Available -Alzheimer’s Disease* -Lou Gehrig’s Disease (amyotrophic lateral sclerosis) -breast/ovarian/colon cancer* -cystic fibrosis -fragile X syndrome -Huntington’s Disease -sickle cell anemia -Tay-Sachs Disease *susceptibility test only

Bioinformatics/PCR Lab Linking Genetics to Multifactorial Disease (greater risk; involves population studies) -cancer -other current avenues of research • peripheral arterial disease • type I diabetes • schizophrenia/bipolar disorder • calcium oxalate stone disease • Chron’s disease • ulcerative colitis

Bioinformatics/PCR Lab How does having a certain genetic marker affect chances of getting brain cancer? How does having a specific nucleotide sequence in a specific region of DNA affect chances of getting brain cancer?

Bioinformatics/PCR Lab How do you determine what the nucleotide sequence is in a piece of DNA? How do you detect differences in nucleotide sequences in a piece of DNA? How do you determine if these nucleotide differences are related to disease risk?
Bioinformatics/PCR Lab Bioinformatics: • human genome project (link in course website) • 1990 -2003 • identify all human genes • sequence human chromosomes • make available to all (databases)
Bioinformatics/PCR Lab Bioinformatics: • human genome project (link in course website) • comparisons of sequence information • within species • across species • other species genomes sequenced “They’re Sequencing a What? ” Science News, 10/9/04, pp. 234 -36.

Bioinformatics/PCR Lab Bioinformatics: • human genome project (link in course website) • comparisons of sequence information • BLAST search results? • specific DNA segment to work with? • “real” interest in this DNA? • why are we using this segment?

Bioinformatics/PCR Lab General Lab Procedures: 1) isolate DNA from hair (“hair digest”) 2) amplify (make many copies of) DNA from hair using PCR 3) cut PCR product with special enzymes • restriction enzymes • “restriction digest”

Bioinformatics/PCR Lab General Lab Procedures: 4) visualize cut PCR product on a gel 5) compare patterns (RFLPs) 6) follow people around for decades to see who (which RFLP) gets brain cancer!
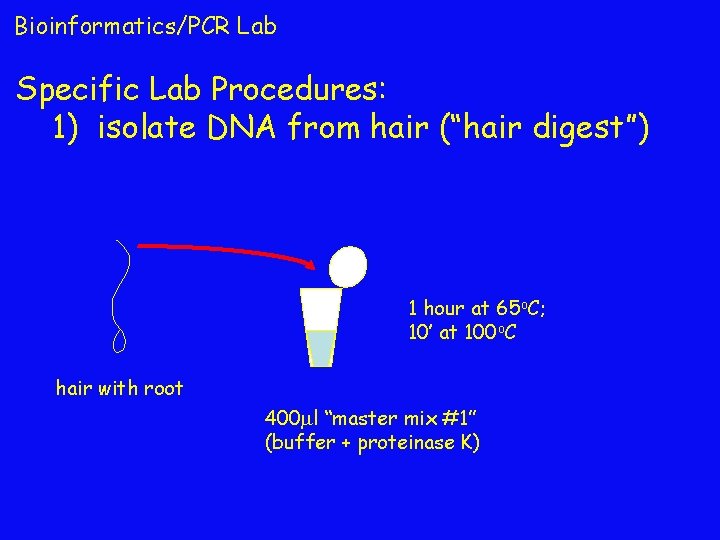
Bioinformatics/PCR Lab Specific Lab Procedures: 1) isolate DNA from hair (“hair digest”) 1 hour at 65 o. C; 10’ at 100 o. C hair with root 400 l “master mix #1” (buffer + proteinase K)
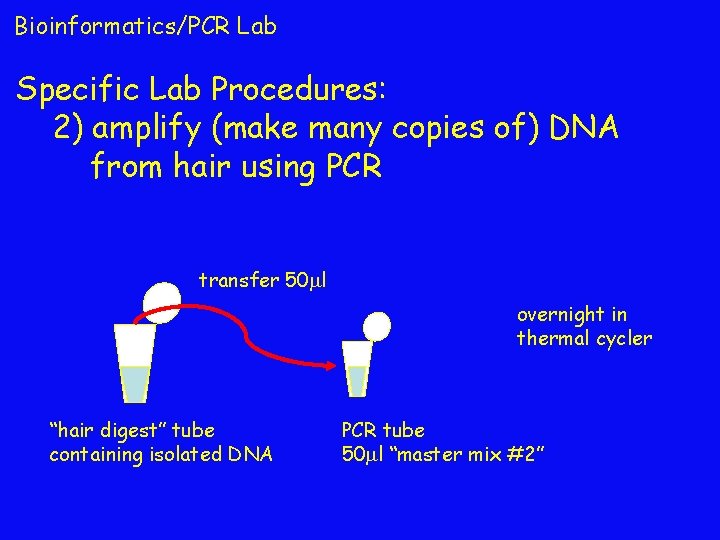
Bioinformatics/PCR Lab Specific Lab Procedures: 2) amplify (make many copies of) DNA from hair using PCR transfer 50 l overnight in thermal cycler “hair digest” tube containing isolated DNA PCR tube 50 l “master mix #2”
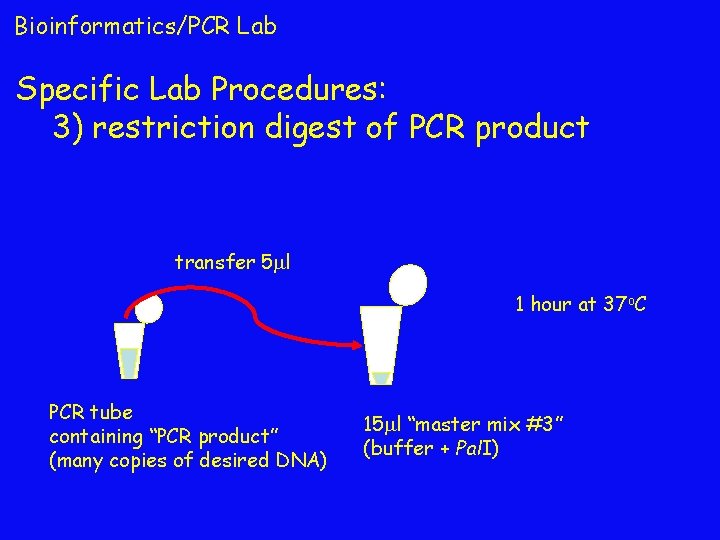
Bioinformatics/PCR Lab Specific Lab Procedures: 3) restriction digest of PCR product transfer 5 l 1 hour at 37 o. C PCR tube containing “PCR product” (many copies of desired DNA) 15 l “master mix #3” (buffer + Pal. I)

Bioinformatics/PCR Lab Specific Lab Procedures: 4) visualize cut PCR product on a gel transfer 5 l transfer 15 l new 1. 5 ml microfuge tube “undigested PCR product” DNA loading dye transfer 5 l “digested PCR product”
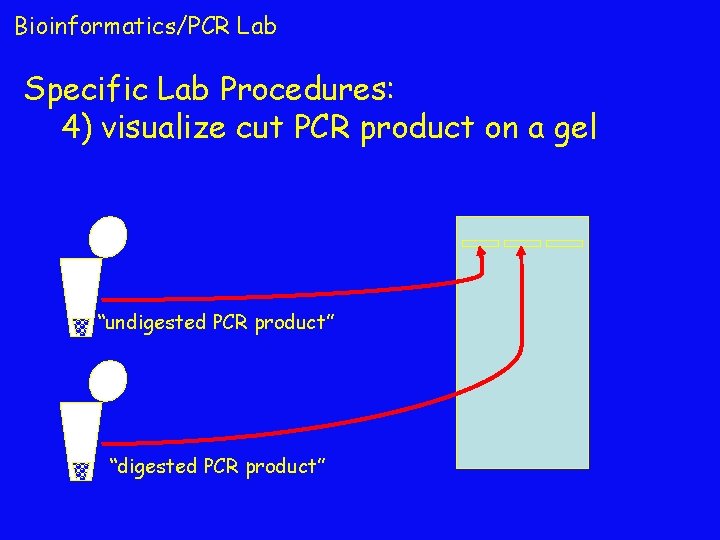
Bioinformatics/PCR Lab Specific Lab Procedures: 4) visualize cut PCR product on a gel “undigested PCR product” “digested PCR product”
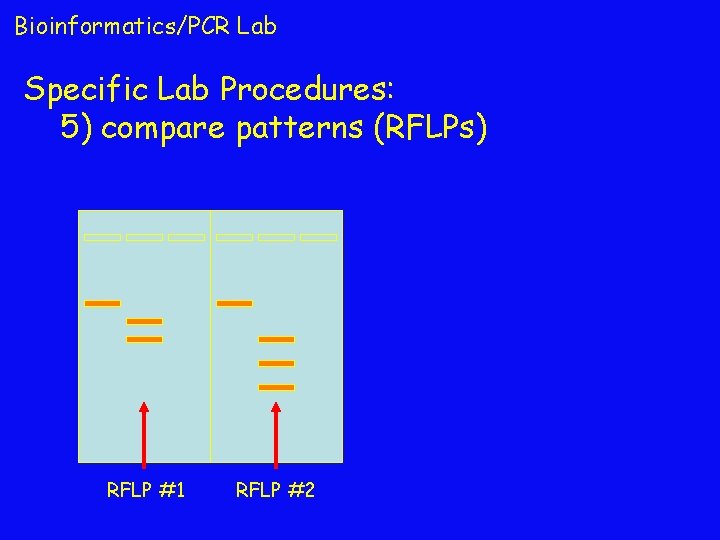
Bioinformatics/PCR Lab Specific Lab Procedures: 5) compare patterns (RFLPs) RFLP #1 RFLP #2
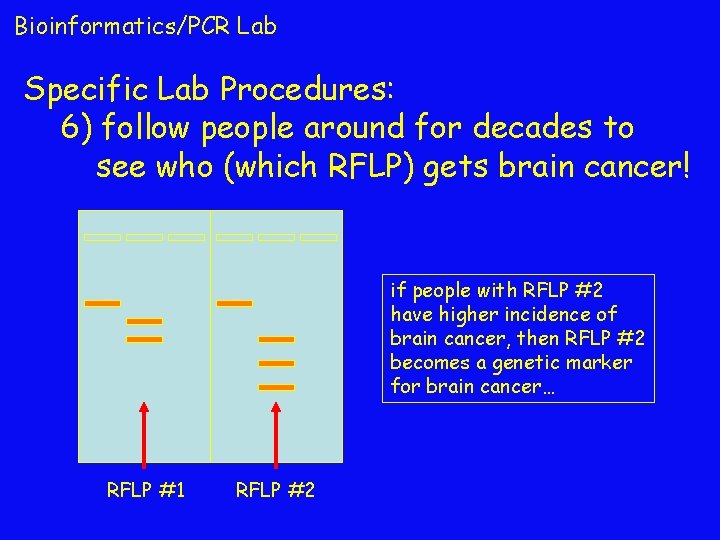
Bioinformatics/PCR Lab Specific Lab Procedures: 6) follow people around for decades to see who (which RFLP) gets brain cancer! if people with RFLP #2 have higher incidence of brain cancer, then RFLP #2 becomes a genetic marker for brain cancer… RFLP #1 RFLP #2

Bioinformatics/PCR Lab Techniques to Know: 1) PCR 2) Restriction Digest 3) Gel Electrophoresis
Bioinformatics/PCR Lab PCR: Polymerase Chain Reaction • method of making many copies (amplifying) of a specific sequence (region) of DNA in vitro (in a test tube) • developed by Kary Mullis, 1983 • Nobel Prize in Chemistry, 1993
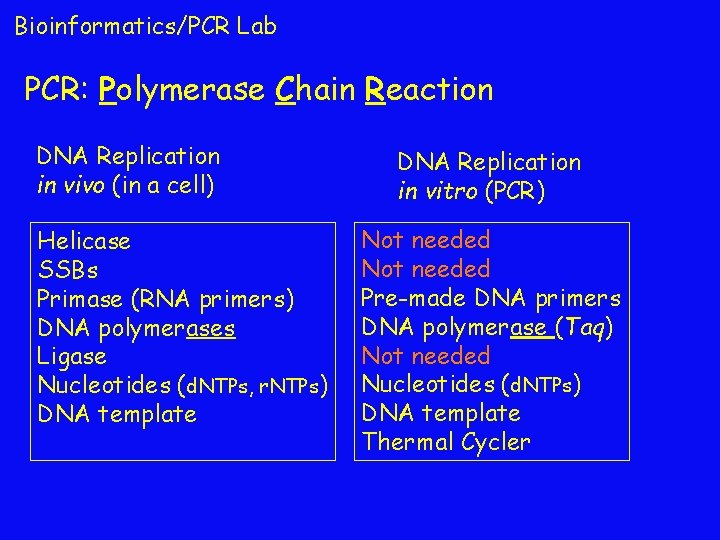
Bioinformatics/PCR Lab PCR: Polymerase Chain Reaction DNA Replication in vivo (in a cell) Helicase SSBs Primase (RNA primers) DNA polymerases Ligase Nucleotides (d. NTPs, r. NTPs) DNA template DNA Replication in vitro (PCR) Not needed Pre-made DNA primers DNA polymerase (Taq) Not needed Nucleotides (d. NTPs) DNA template Thermal Cycler
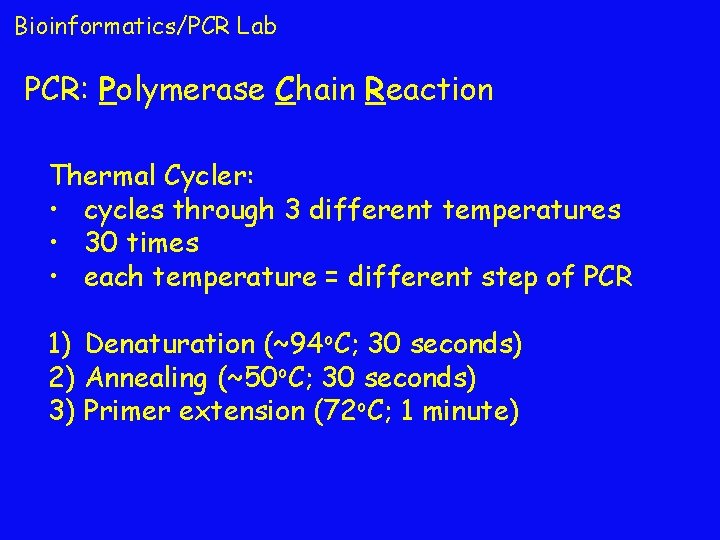
Bioinformatics/PCR Lab PCR: Polymerase Chain Reaction Thermal Cycler: • cycles through 3 different temperatures • 30 times • each temperature = different step of PCR 1) Denaturation (~94 o. C; 30 seconds) 2) Annealing (~50 o. C; 30 seconds) 3) Primer extension (72 o. C; 1 minute)
Bioinformatics/PCR Lab PCR: Polymerase Chain Reaction 1) Denaturation (~94 o. C; 30 seconds) • ds. DNA template separates into two ss. DNA • cellular function: helicase, SSBs
Bioinformatics/PCR Lab PCR: Polymerase Chain Reaction 1) Denaturation (~94 o. C; 30 seconds) 2) Annealing (~50 o. C; 30 seconds) • pre-made DNA primers H-bond to template • • • define region to be amplified one primer for each strand of template orientation correct with respect to 3’/5’ • cellular function: primase, primer removal, ligase
Bioinformatics/PCR Lab PCR: Polymerase Chain Reaction 1) Denaturation (~94 o. C; 30 seconds) 2) Annealing (~50 o. C; 30 seconds) 3) Primer extension (72 o. C; 1 minute) • Taq DNA polymerase adds nucleotides to 3’OH • starts from primer • cellular function: same
Bioinformatics/PCR Lab PCR: Polymerase Chain Reaction • In the olden days… • 3 waterbaths at each temperature • Fresh DNA polymerase added each cycle • Automation • Thermal cycler controls temperature • Taq DNA polymerase remains stable • Taq = Thermus aquaticus

Bioinformatics/PCR Lab PCR: Polymerase Chain Reaction Animation: www. dnalc. org/shockwave/pcranwhole. html

Bioinformatics/PCR Lab Restriction Digest • Restriction enzymes • Restriction digest = fragments of certain length due to specific sequences

Bioinformatics/PCR Lab Restriction Digest Recognizes “the” and cuts between h - e themonkeyandthedonkeyaretherenow (1) th emonkeyandth edonkeyareth erenow (4) themonkeyandthedonkeyarethreenow (1) th emonkeyandth edonkeyarethreenow (3)

Bioinformatics/PCR Lab Gel Electrophoresis Separation of molecules based on their physical/chemical properties through a matrix by applying an electric current

Bioinformatics/PCR Lab Gel Electrophoresis Separation of molecules nucleic acids, proteins based on their physical/chemical properties through a matrix by applying an electric current

Bioinformatics/PCR Lab Gel Electrophoresis Separation of molecules nucleic acids, proteins based on their physical/chemical properties size, charge through a matrix by applying an electric current
Bioinformatics/PCR Lab Gel Electrophoresis Separation of molecules nucleic acids, proteins based on their physical/chemical properties size, charge through a matrix agarose, polyacrylamide by applying an electric current

Bioinformatics/PCR Lab Gel Electrophoresis Separation of molecules nucleic acids (DNA) based on their physical/chemical properties size (length in base pairs - bp) through a matrix agarose by applying an electric current
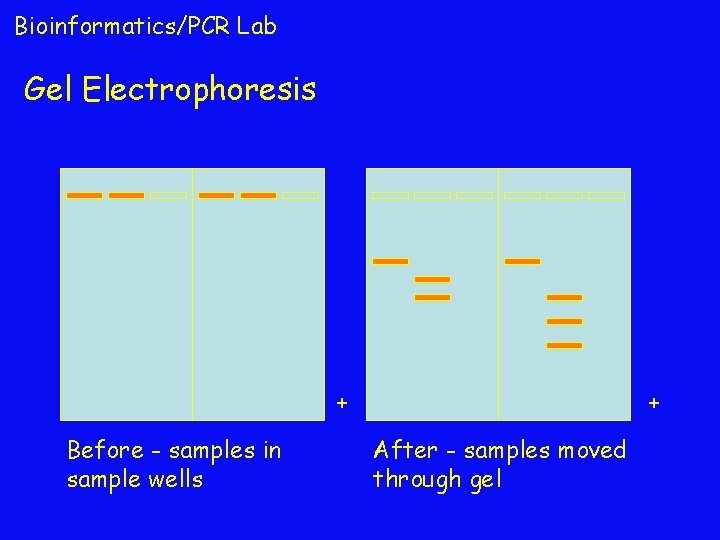
Bioinformatics/PCR Lab Gel Electrophoresis + Before - samples in sample wells + After - samples moved through gel
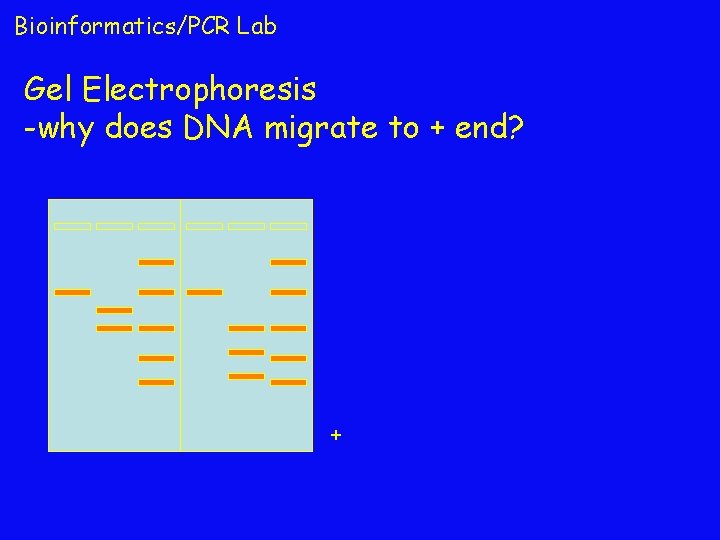
Bioinformatics/PCR Lab Gel Electrophoresis -why does DNA migrate to + end? +
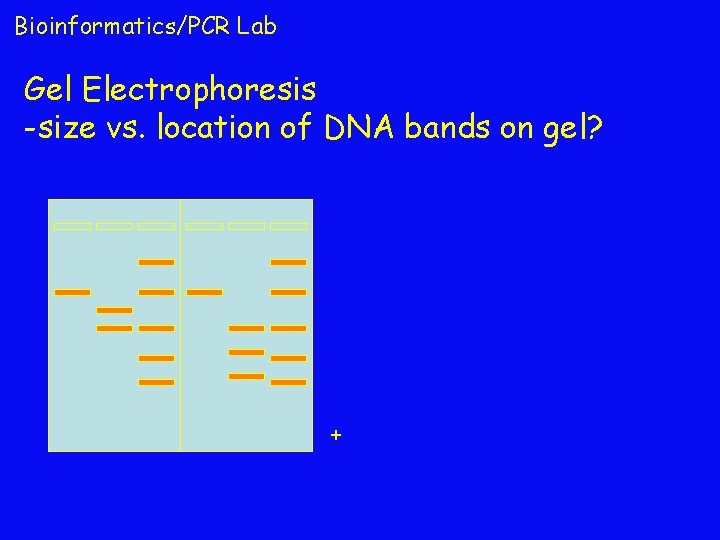
Bioinformatics/PCR Lab Gel Electrophoresis -size vs. location of DNA bands on gel? +
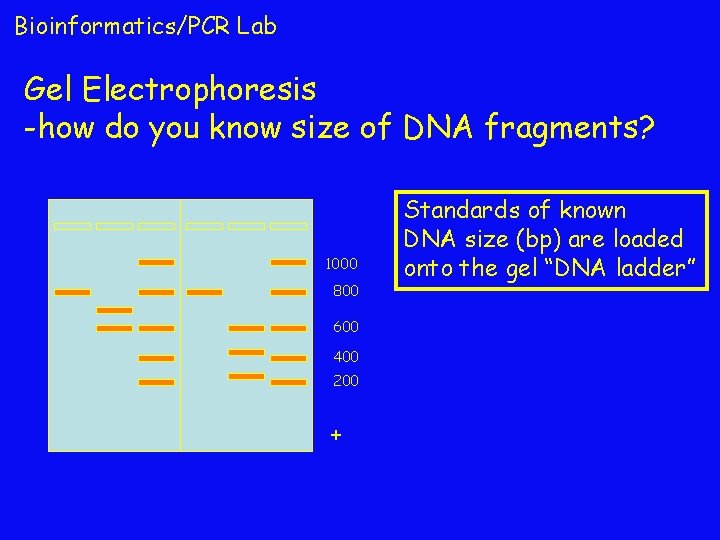
Bioinformatics/PCR Lab Gel Electrophoresis -how do you know size of DNA fragments? 1000 800 600 400 200 + Standards of known DNA size (bp) are loaded onto the gel “DNA ladder”
- Slides: 39