Bioinformatics Richard Tseng and Ishawar Hosamani Outline Homology

Bioinformatics Richard Tseng and Ishawar Hosamani
Outline • Homology modeling (Ishwar) • Structural analysis – Structure prediction – Structure comparisons • Cluster analysis – Partitioning method – Density-based method • Phylogenetic analaysis
Structural Analysis • Overview – Structure prediction – Structural alignment – Similarity
• Tools for protein structure prediction – Protein • Secondary structure prediction: SSEA http: //protein. cribi. unipd. it/ssea/ • Tertiary structure prediction: – Wurst: http: //www. zbh. uni-hamburg. de/wurst/ – LOOPP: http: //cbsuapps. tc. cornell. edu/loopp. aspx

• WURST( Torda et al. (2004) Wurst: A protein threading server with a structural scoring function, sequence profiles and optimized substitution matrices Nucleic Acids Res. , 32, W 532 -W 535 ) • Rationale – Alignment: Sequence to structure alignments are done with a Smith-Waterman style alignment and the Gotoh algorithm – Score function: fragment-based sequence to structure compatibility score and a pure sequence -sequence component substitution score – Library: Dali PDB 90 (24599 srtuctures)
• Tools for structure comparison – Pair structures comparison: • Top. Match • Matras: (http: //biunit. naist. jp/matras/) – Multiple structures comparison: • 3 D-surfer • Matras: (http: //biunit. naist. jp/matras/)
• Top. Match (Sippl & Wiederstein (2008) A note on difficult structure alignment problems. Bioinformatics 24, 426 -427) – Rationale: • Structure alignment: http: //www. cgl. ucsf. edu/home/meng/grpmt/structalign. html • Similarity measurement – Input format • PDB, SCOP and CATH code • PDB structure directly – Exercise: http: //topmatch. services. came. sbg. ac. at/

• 3 D-surfer (David La et al. 3 D-SURFER: software for high throughput protein surface comparison and analysis. Bioinformatics , in press. (2009)) – Rationale 1. Define a surface function 2. Transform the surface function into a 3 D Zernike description function – Input format • PDB and CATH code • PDB structure directly – Exercise: http: //dragon. bio. purdue. edu/3 d-surfer/
Cluster analysis • Goal: – Grouping the data into classes or clusters, so that objects within a cluster have high similarity in comparison to one another but are very dissimilar to objects in other clusters. • Methods – Partitioning method: k-means – Density-based method: Ordering Points to Identify the Clustering Structure (OPTICS)

• k-means – Rationale: Partition n observations into k clusters in which each observation belongs to the cluster with the nearest mean – Exercise http: //cgm. cs. ntust. edu. tw/etrex/k. Means. Clustering 2. html
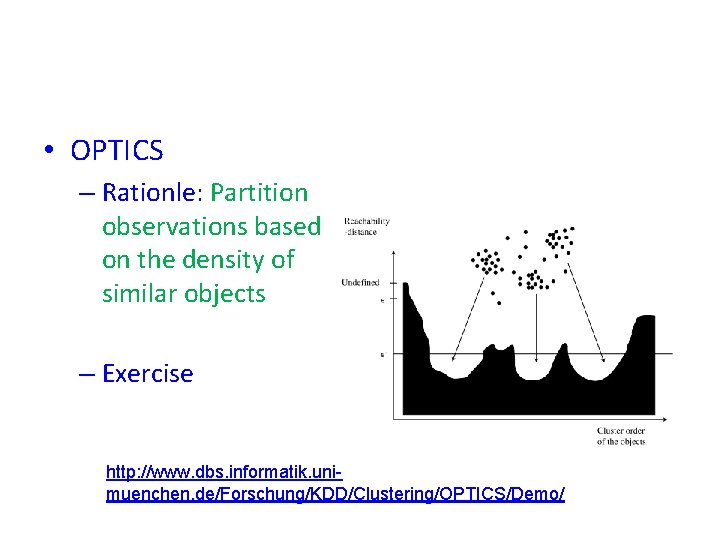
• OPTICS – Rationle: Partition observations based on the density of similar objects – Exercise http: //www. dbs. informatik. unimuenchen. de/Forschung/KDD/Clustering/OPTICS/Demo/
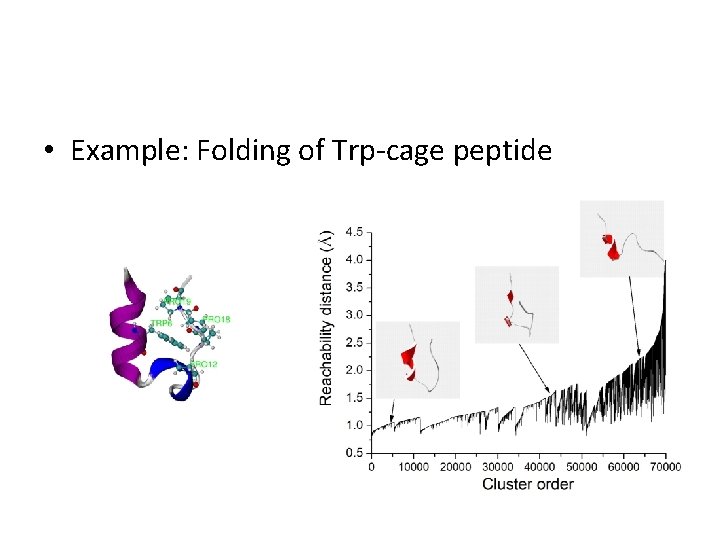
• Example: Folding of Trp-cage peptide
Phylogenetic analysis • Overviews – Comparisons of more than two sequences – Analysis of gene families, including functional predictions – Estimation of evolutionary relationships among organisms

• Theoretical tree – Parsimony method – Distance matrix method – Maximum likelihood and Bayesian method – Invariants method

• Software – Collections of tools http: //evolution. genetics. washington. edu/phylip/software. html – A web server version for tree construction and display • PHYLIP, http: //bioweb 2. pasteur. fr/phylogeny/intro-en. html • Interactive tree of life, http: //itol. embl. de/ – Mostly common used stand alone software • PHYLIP, tool for evaluating similarity of nucleotide and amino acid sequences. http: //evolution. gs. washington. edu/phylip. html • Tree. View, tool for visualization and manipulation of family tree. http: //taxonomy. zoology. gla. ac. uk/rod/treeview. html • Matlab - bioinformatics tool box
• Example: Alignment phylogenetic tree of Tubulin family – Searching homologous sequences of Tubulin (PDB code: 1 JFF) from RCSB protein databank • Blast for pair sequence alignment • Clustalw for comparative sequence alignment – Evaluating protein distance matrix • using “Protdist” of PHYILIP (Particularly, Point Accepted Mutation (PAM) matrix is used) – Clustering proteins using “Neighbor” of PHYILIP (Neightboring-Joint method is considered)
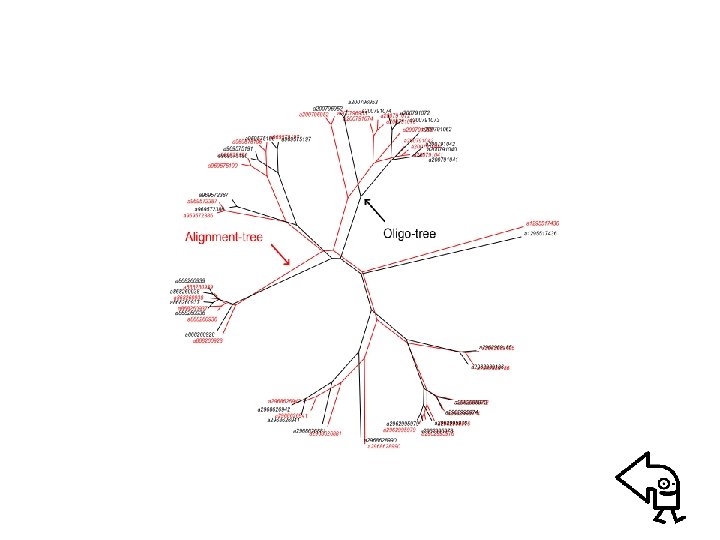
• Example: n-distance phylogenetic tree – Evaluating n-distance matrix • n-distance method – Clustering proteins using “Neighbor” of PHYILIP (Neightboring-Joint method is considered) • 16 S and 18 S Ribosomal RNA sequenecs of 35 organisms
Summary • Homology modeling • Tools for structure prediction and comparisons • Tools for phylogenetic tree construction Thanks for your attention!!
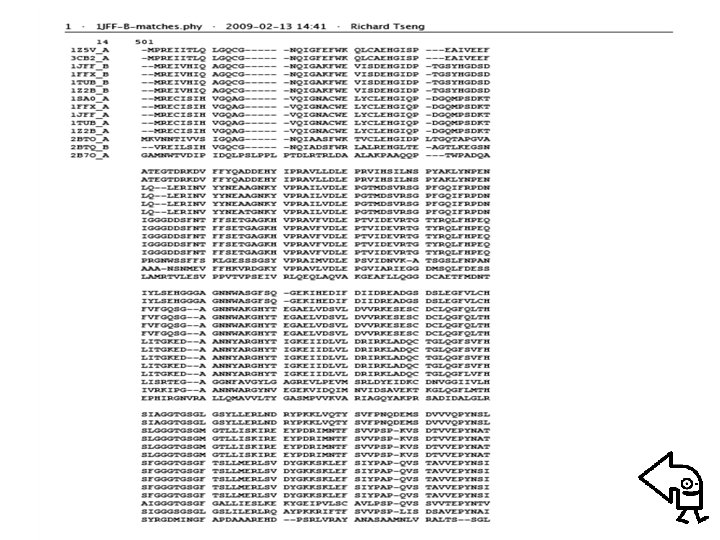
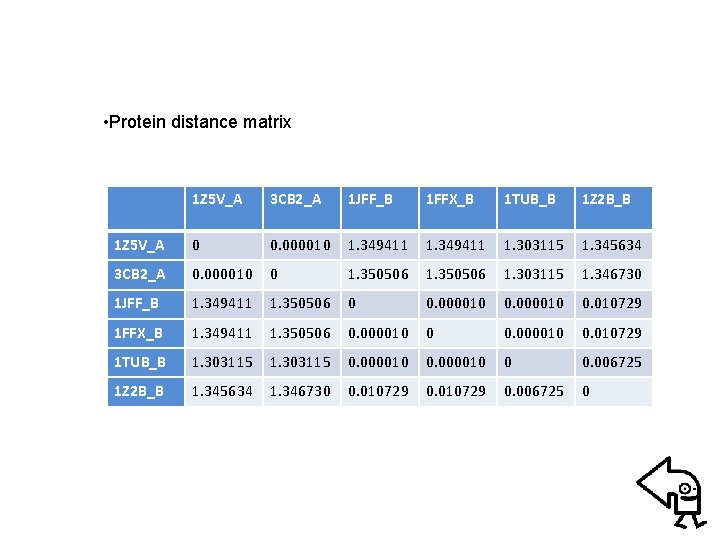
• Protein distance matrix 1 Z 5 V_A 3 CB 2_A 1 JFF_B 1 FFX_B 1 TUB_B 1 Z 2 B_B 1 Z 5 V_A 0 0. 000010 1. 349411 1. 303115 1. 345634 3 CB 2_A 0. 000010 0 1. 350506 1. 303115 1. 346730 1 JFF_B 1. 349411 1. 350506 0 0. 000010 0. 010729 1 FFX_B 1. 349411 1. 350506 0. 000010 0. 010729 1 TUB_B 1. 303115 0. 000010 0 0. 006725 1 Z 2 B_B 1. 345634 1. 346730 0. 010729 0. 006725 0
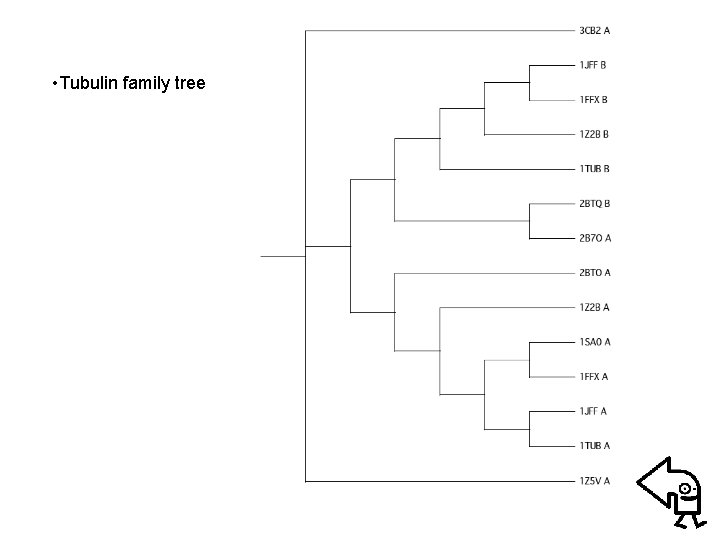
• Tubulin family tree

• n-distance method – Frequency count of “n-letter words” MREIVHIQAGQCGNQIGAKFWEVISDEHGIDPTGSYHGDSDLQLERINVYYNE – n-dsiatnce matrix – Advantage: 1. Identify fully conservative words located at nearly the same sites 2. Effecient
- Slides: 24