Bioinformatics and Protein Structural Analysis The molecular structures
Bioinformatics and Protein Structural Analysis The molecular structures of proteins are complex and can be defined at various levels. These structures can also be predicted from their amino-acid sequences. Protein structure prediction is one of the most widespread fields of research in bioinformatics. Surabhi Agarwal
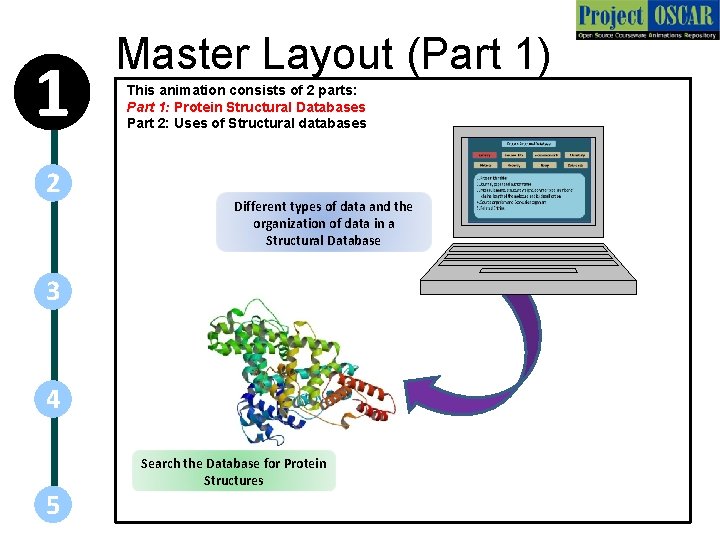
1 2 Master Layout (Part 1) This animation consists of 2 parts: Part 1: Protein Structural Databases Part 2: Uses of Structural databases Different types of data and the organization of data in a Structural Database 3 4 5 Search the Database for Protein Structures
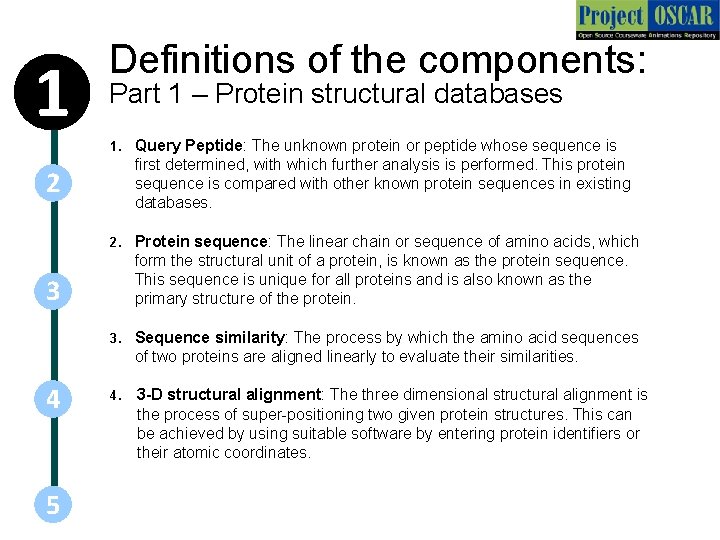
1 Definitions of the components: Part 1 – Protein structural databases 1. Query Peptide: The unknown protein or peptide whose sequence is first determined, with which further analysis is performed. This protein sequence is compared with other known protein sequences in existing databases. 2. Protein sequence: The linear chain or sequence of amino acids, which form the structural unit of a protein, is known as the protein sequence. This sequence is unique for all proteins and is also known as the primary structure of the protein. 3. Sequence similarity: The process by which the amino acid sequences of two proteins are aligned linearly to evaluate their similarities. 4. 3 -D structural alignment: The three dimensional structural alignment is the process of super-positioning two given protein structures. This can be achieved by using suitable software by entering protein identifiers or their atomic coordinates. 2 3 4 5
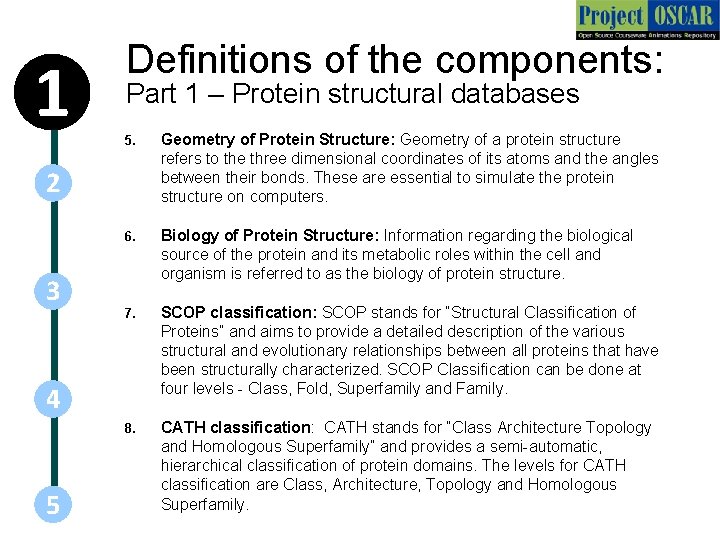
1 Definitions of the components: Part 1 – Protein structural databases 5. Geometry of Protein Structure: Geometry of a protein structure refers to the three dimensional coordinates of its atoms and the angles between their bonds. These are essential to simulate the protein structure on computers. 6. Biology of Protein Structure: Information regarding the biological source of the protein and its metabolic roles within the cell and organism is referred to as the biology of protein structure. 7. SCOP classification: SCOP stands for “Structural Classification of Proteins” and aims to provide a detailed description of the various structural and evolutionary relationships between all proteins that have been structurally characterized. SCOP Classification can be done at four levels - Class, Fold, Superfamily and Family. 8. CATH classification: CATH stands for “Class Architecture Topology and Homologous Superfamily” and provides a semi-automatic, hierarchical classification of protein domains. The levels for CATH classification are Class, Architecture, Topology and Homologous Superfamily. 2 3 4 5
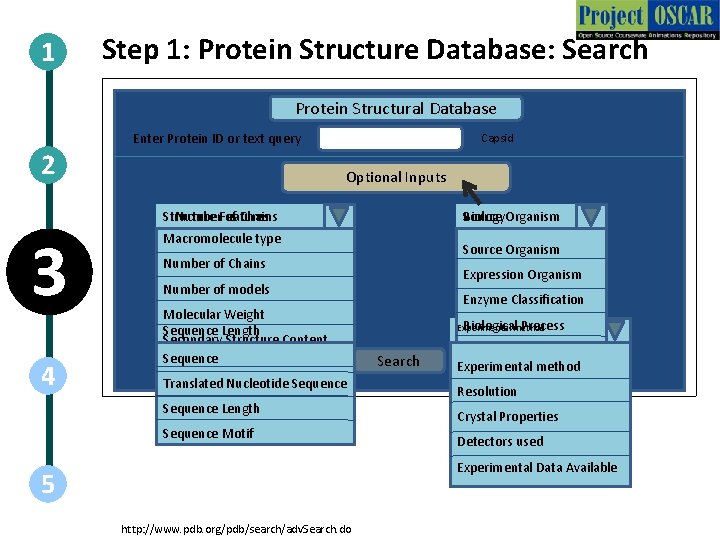
1 Step 1: Protein Structure Database: Search Protein Structural Database Enter Protein ID or text query 2 Capsid Optional Inputs Structure Number. Features of Chains 3 4 Biology. Organism Source Macromolecule type Retro Transcribing Source Organism. Viruses 10 Number of Chains Expression Organism Number of models Molecular Weight Sequence Length Features Secondary Structure Content Sequence < 500 Secondary Structure Length Translated Nucleotide Sequence SCOP classification Sequence Length CATH classification Sequence Motif Enzyme Classification Biologicalmethod Process Experimental Experiment Search Cellular component X-RAY CRYSTALLOGRAPHY Experimental method Resolution Crystal Properties Detectors used Experimental Data Available 5 http: //www. pdb. org/pdb/search/adv. Search. do
1 2 3 Step 1: Protein Structure Database: Search Action Description of the action Schematic for Database functioning Follow the steps as shown in the animations. First show the basic layout of the database. Then input the test “Capsid” in the text box on the top of the page. For each 4 categories, when the down-link gets clicked announce the options as the mouse hovers on them. The downlink in the animation should look like the downlink in web-pages. Recreate all images. 4 5 http: //www. pdb. org/pdb/search/adv. Search. do Audio Narration The protein structural databases contain a basic search box which requires the input for an identifier of the protein. This identifier can be the protein name, key-word, ID, author, etc. In this example, we take the case of Viral Capsid Proteins. These databases have advanced search features which are optional but help in making the query very specific. The general options can be categorized in 4 broad classes. Structural Features, Biology, Sequence Data and Experimental Details.
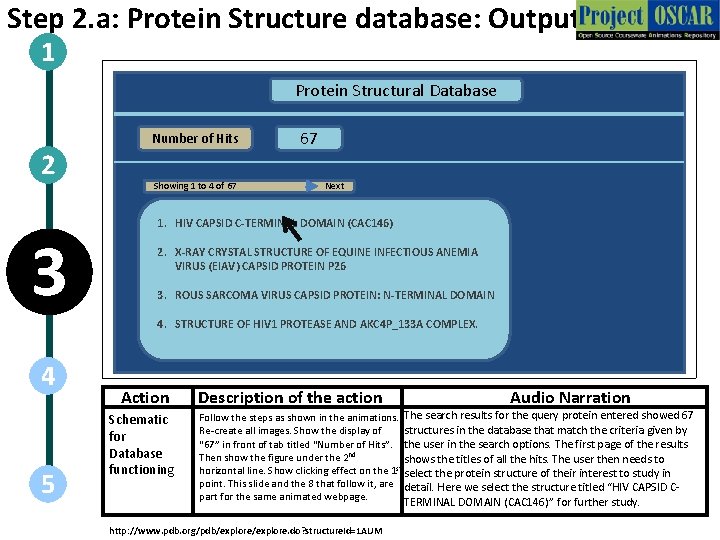
Step 2. a: Protein Structure database: Output 1 Protein Structural Database Number of Hits 2 3 Showing 1 to 4 of 67 67 Next 1. HIV CAPSID C-TERMINAL DOMAIN (CAC 146) 2. X-RAY CRYSTAL STRUCTURE OF EQUINE INFECTIOUS ANEMIA VIRUS (EIAV) CAPSID PROTEIN P 26 3. ROUS SARCOMA VIRUS CAPSID PROTEIN: N-TERMINAL DOMAIN 4. STRUCTURE OF HIV 1 PROTEASE AND AKC 4 P_133 A COMPLEX. 4 5 Action Schematic for Database functioning Description of the action Audio Narration Follow the steps as shown in the animations. The search results for the query protein entered showed 67 structures in the database that match the criteria given by Re-create all images. Show the display of “ 67” in front of tab titled “Number of Hits”. the user in the search options. The first page of the results Then show the figure under the 2 nd shows the titles of all the hits. The user then needs to horizontal line. Show clicking effect on the 1 st select the protein structure of their interest to study in point. This slide and the 8 that follow it, are detail. Here we select the structure titled “HIV CAPSID Cpart for the same animated webpage. TERMINAL DOMAIN (CAC 146)” for further study. http: //www. pdb. org/pdb/explore. do? structure. Id=1 AUM
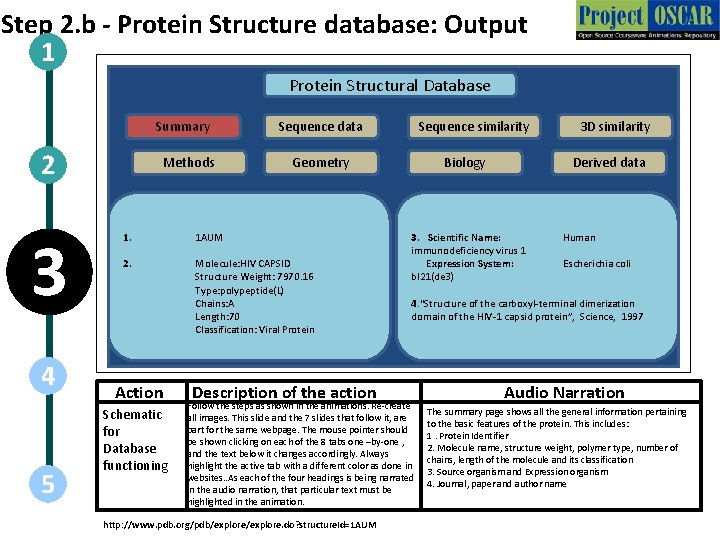
Step 2. b - Protein Structure database: Output 1 Protein Structural Database Summary 2 3 4 5 Methods Sequence data Geometry 1. 1 AUM 2. Molecule: HIV CAPSID Structure Weight: 7970. 16 Type: polypeptide(L) Chains: A Length: 70 Classification: Viral Protein Action Schematic for Database functioning Sequence similarity Description of the action Biology Derived data 3. Scientific Name: immunodeficiency virus 1 Expression System: bl 21(de 3) Human Escherichia coli 4. “Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein”, Science, 1997 Follow the steps as shown in the animations. Re-create all images. This slide and the 7 slides that follow it, are part for the same webpage. The mouse pointer should be shown clicking on each of the 8 tabs one –by-one , and the text below it changes accordingly. Always highlight the active tab with a different color as done in websites. . As each of the four headings is being narrated in the audio narration, that particular text must be highlighted in the animation. http: //www. pdb. org/pdb/explore. do? structure. Id=1 AUM 3 D similarity Audio Narration The summary page shows all the general information pertaining to the basic features of the protein. This includes: 1. Protein Identifier 2. Molecule name, structure weight, polymer type, number of chains, length of the molecule and its classification 3. Source organism and Expression organism 4. Journal, paper and author name
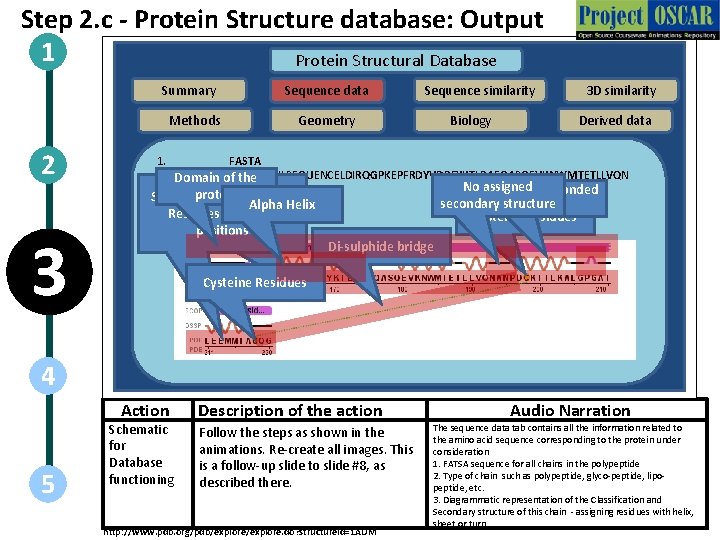
Step 2. c - Protein Structure database: Output 1 Protein Structural Database Summary Methods 2 3 Sequence data Sequence similarity Geometry Biology 3 D similarity Derived data 1. FASTA >1 AUM: A|PDBID|CHAIN|SEQUENCELDIRQGPKEPFRDYVDRFYKTLRAEQASQEVKNWMTETLLVQN Domain of the No assigned ANPDCKTILKALGPGATLEEMMTACQG Hydrogen Bonded protein Sequence of Amino Acid Alpha Helix andpolypeptide(L) their 2. Residues Chain Type: positions 3. secondary structure Turn Cysteine Residues Di-sulphide bridge Cysteine Residues 4 Action 5 Schematic for Database functioning Description of the action Follow the steps as shown in the animations. Re-create all images. This is a follow-up slide to slide #8, as described there. http: //www. pdb. org/pdb/explore. do? structure. Id=1 AUM Audio Narration The sequence data tab contains all the information related to the amino acid sequence corresponding to the protein under consideration 1. FATSA sequence for all chains in the polypeptide 2. Type of chain such as polypeptide, glyco-peptide, lipopeptide, etc. 3. Diagrammatic representation of the Classification and Secondary structure of this chain - assigning residues with helix, sheet or turn
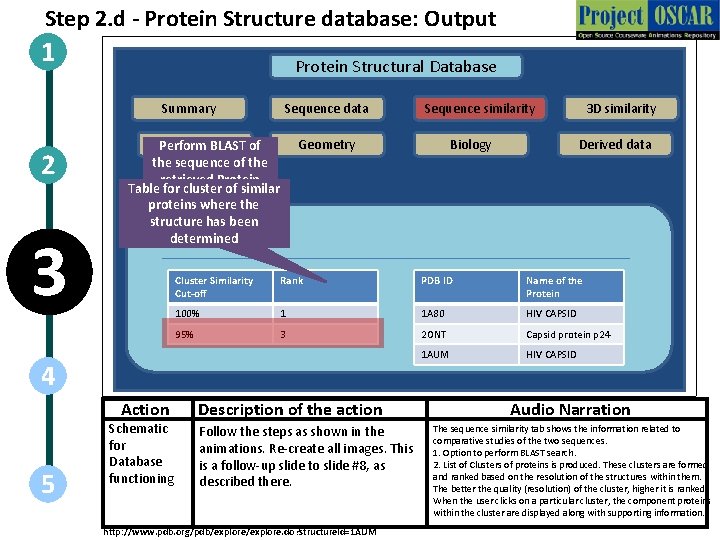
Step 2. d - Protein Structure database: Output 1 Protein Structural Database Summary 2 3 Sequence data Methods Perform BLAST of the sequence of the retrieved Protein Table for cluster of similar proteins where the structure has been determined BLAST Geometry 5 Schematic for Database functioning Biology 3 D similarity Derived data Cluster Similarity Cut-off Rank PDB ID Name of the Protein 100% 1 1 A 80 HIV CAPSID 95% 3 2 ONT Capsid protein p 24 1 AUM HIV CAPSID 4 Action Sequence similarity Description of the action Follow the steps as shown in the animations. Re-create all images. This is a follow-up slide to slide #8, as described there. http: //www. pdb. org/pdb/explore. do? structure. Id=1 AUM Audio Narration The sequence similarity tab shows the information related to comparative studies of the two sequences. 1. Option to perform BLAST search. 2. List of Clusters of proteins is produced. These clusters are formed and ranked based on the resolution of the structures within them. The better the quality (resolution) of the cluster, higher it is ranked. When the user clicks on a particular cluster, the component proteins within the cluster are displayed along with supporting information. .
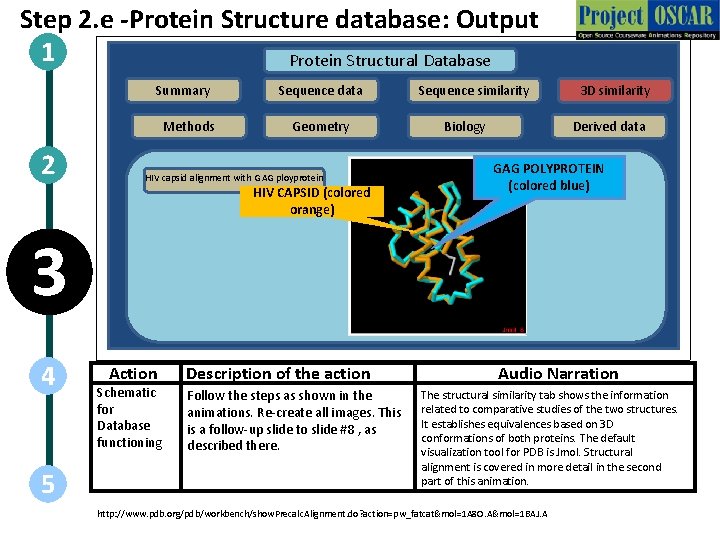
Step 2. e -Protein Structure database: Output 1 Protein Structural Database Summary Methods 2 Sequence data Geometry HIV capsid alignment with GAG ployprotein HIV CAPSID (colored orange) Sequence similarity Biology 3 D similarity Derived data GAG POLYPROTEIN (colored blue) 3 4 5 Action Schematic for Database functioning Description of the action Follow the steps as shown in the animations. Re-create all images. This is a follow-up slide to slide #8 , as described there. Audio Narration The structural similarity tab shows the information related to comparative studies of the two structures. It establishes equivalences based on 3 D conformations of both proteins. The default visualization tool for PDB is Jmol. Structural alignment is covered in more detail in the second part of this animation. http: //www. pdb. org/pdb/workbench/show. Precalc. Alignment. do? action=pw_fatcat&mol=1 A 8 O. A&mol=1 BAJ. A
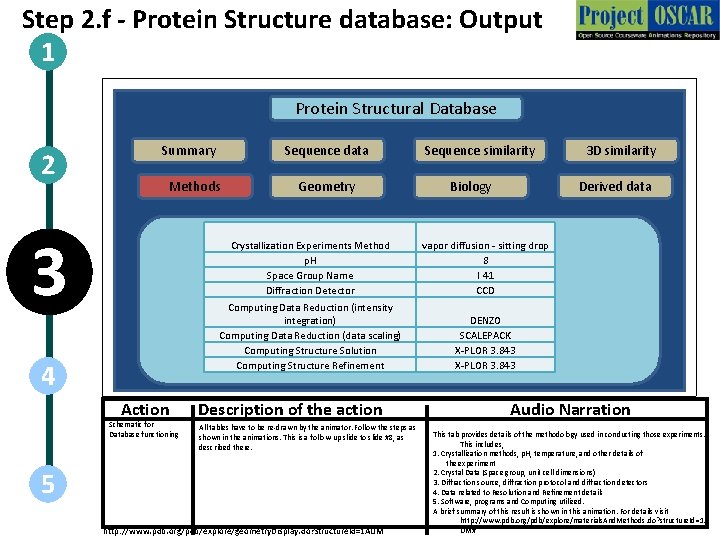
Step 2. f - Protein Structure database: Output 1 Protein Structural Database 2 Summary Sequence data Methods 3 4 Action Schematic for Database functioning Geometry Sequence similarity Biology Derived data Crystallization Experiments Method p. H Space Group Name Diffraction Detector vapor diffusion - sitting drop 8 I 41 CCD Computing Data Reduction (intensity integration) Computing Data Reduction (data scaling) Computing Structure Solution Computing Structure Refinement DENZO SCALEPACK X-PLOR 3. 843 Description of the action All tables have to be re-drawn by the animator. Follow the steps as shown in the animations. This is a follow-up slide to slide #8, as described there. 5 http: //www. pdb. org/pdb/explore/geometry. Display. do? structure. Id=1 AUM 3 D similarity Audio Narration This tab provides details of the methodology used in conducting those experiments. This includes, 1. Crystallization methods, p. H, temperature, and other details of the experiment 2. Crystal Data (Space group, unit cell dimensions) 3. Diffraction source, diffraction protocol and diffraction detectors 4. Data related to Resolution and Refinement details 5. Software, programs and Computing utilized. A brief summary of this result is shown in this animation. For details visit http: //www. pdb. org/pdb/explore/materials. And. Methods. do? structure. Id=1 A UM#
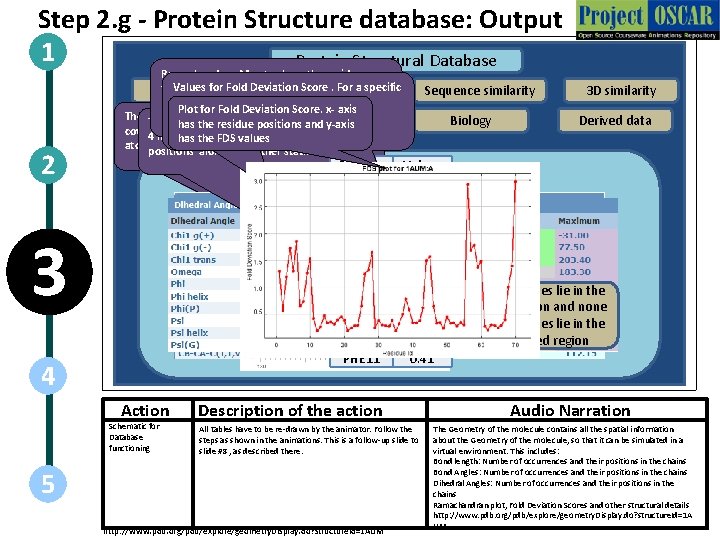
Step 2. g - Protein Structure database: Output 1 Protein Structural Database 2 Ramachandran Map to show the residues Values Deviation Score. For a specific that lie in for the. Fold favored region (outlined in Dark Summary Sequence data reference FDS isregion a multiple of thein Blue) and thevalue, permitted (outlined Plot for Fold Deviation Score. x- axis standard deviation The position, total number, range of the planesinof blue) Methods Geometry Thelight angle formed by 32 positions consecutive has the residue andatoms y-axis covalent bond lengths between two adjacent native conformation of a protein and their 4 linearly bonded Their occurrence, has the FDS atoms. values atoms in a protein molecule statistics positions along with other statistics. Residue LEU 1 ASP 2 ILE 3 ARG 4 GLN 5 GLY 6 PRO 7 LYS 8 GLU 9 PRO 10 PHE 11 3 4 Action Schematic for Database functioning Sequence similarity Biology Values 1. 29 0. 56 1. 19 1. 73 1. 29 1. 85 0. 65 0. 73 1. 27 1. 53 0. 41 Description of the action All tables have to be re-drawn by the animator. Follow the steps as shown in the animations. This is a follow-up slide to slide #8 , as described there. 5 http: //www. pdb. org/pdb/explore/geometry. Display. do? structure. Id=1 AUM 3 D similarity Derived data 67/68 residues lie in the favored region and none of the residues lie in the dis-allowed region Audio Narration The Geometry of the molecule contains all the spatial information about the Geometry of the molecule, so that it can be simulated in a virtual environment. This includes: Bond length: Number of occurrences and their positions in the chains Bond Angles: Number of occurrences and their positions in the chains Dihedral Angles: Number of occurrences and their positions in the chains Ramachandran plot, Fold Deviation Scores and other structural details http: //www. pdb. org/pdb/explore/geometry. Display. do? structure. Id=1 A UM
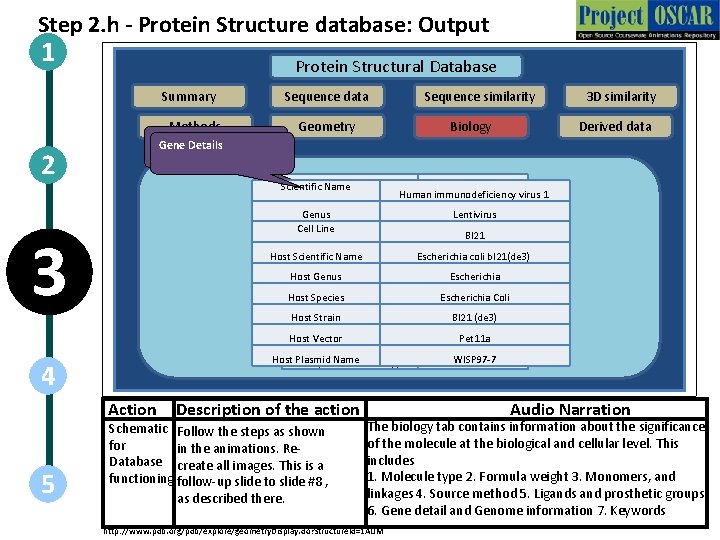
Step 2. h - Protein Structure database: Output 1 Protein Structural Database Summary Methods 2 Protein Details Gene Details Sequence similarity Geometry Biology Description Scientific Name HIV CAPSID Human immunodeficiency virus 1 Genus Fragment Cell Line Nonstandard Linkage Host Scientific Name Nonstandard Monomers Polymer Type Host Genus Formula Weight Host Species 3 Source Method Host Strain Entity Name Host Vector SWS/UNP ID Host Plasmid Name SWS/UNP Accession(s) 4 Action 5 Sequence data Description of the action Schematic Follow the steps as shown for in the animations. Re. Database create all images. This is a functioning follow-up slide to slide #8 , as described there. 3 D similarity Derived data C-TERMINAL DOMAIN, Lentivirus RESIDUES 146 - 231 Bl 21 no Escherichiano coli bl 21(de 3) polypeptide(L) Escherichia 7970. 2 Escherichia Coli genetically manipulated Bl 21 (de 3) CAC 146 Pet 11 a POL_HV 1 N 5 WISP 97 -7 P 12497 Audio Narration The biology tab contains information about the significance of the molecule at the biological and cellular level. This includes 1. Molecule type 2. Formula weight 3. Monomers, and linkages 4. Source method 5. Ligands and prosthetic groups 6. Gene detail and Genome information 7. Keywords http: //www. pdb. org/pdb/explore/geometry. Display. do? structure. Id=1 AUM
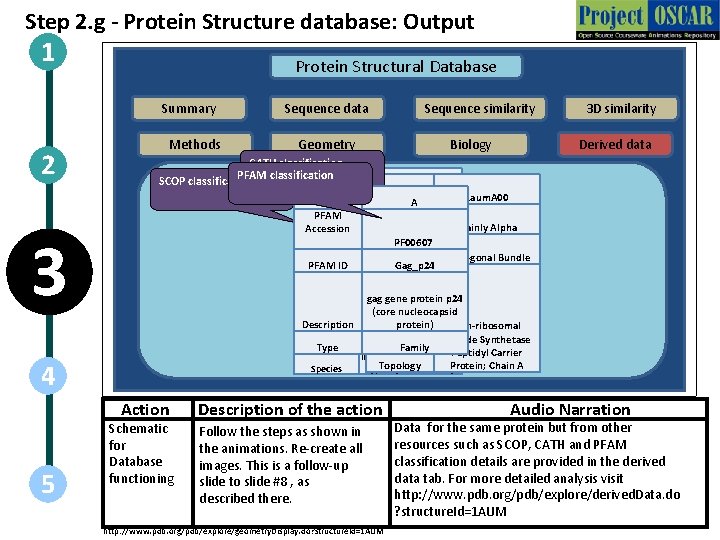
Step 2. g - Protein Structure database: Output 1 Protein Structural Database Summary 2 Methods Sequence data Geometry Biology CATH classification PFAM Domain Info SCOP classification 3 D similarity Derived data d 1 auma_ Class All alpha proteins Domain 1 aum. A 00 Chain A Fold Acyl carrier protein PFAM capsid Class Mainly Alpha Accession Retrovirus dimerization domain. PF 00607 Super-Family like Architecture Orthogonal Bundle capsid PFAM ID Retrovirus Gag_p 24 3 Family Domain Description Type 4 Species Action 5 Sequence similarity Schematic for Database functioning protein C-terminal domain gag gene protein p 24 HIV capsid protein, (core nucleocapsid dimerisation domain protein) Non-ribosomal Peptide Synthetase Human Family Peptidyl Carrier immunodeficiency virus Topology type 1 [Tax. Id: 11676]Protein; Chain A Description of the action Follow the steps as shown in the animations. Re-create all images. This is a follow-up slide to slide #8 , as described there. http: //www. pdb. org/pdb/explore/geometry. Display. do? structure. Id=1 AUM Audio Narration Data for the same protein but from other resources such as SCOP, CATH and PFAM classification details are provided in the derived data tab. For more detailed analysis visit http: //www. pdb. org/pdb/explore/derived. Data. do ? structure. Id=1 AUM
1 Master Layout (Part 2) This animation consists of 2 parts: Part 1: Protein Structural Databases Part 2: Uses of Structural databases 2 3 Protein Structural alignment Secondary Structure Prediction 4 Functional Annotation 5
1 2 3 4 5 Definitions of the components Part 2 – Uses of structural databases 1. Protein Structural Alignment: The geometry of two given protein structures can be compared by means of available software tools that analyse their three dimensional similarity to each other. 2. Protein Structure Prediction: The prospective secondary structures of peptides or proteins can be predicted from a given stretch of amino acid residues by using machine learning algorithms. 3. Machine Learning Algorithms: These are computer algorithms that can be trained from a given classified dataset. Thereafter, these programs train their parameters in a such a way, that they can classify new data. Most widely used Machine Learning Algorithms in Bioinformatics are Artificial Neural Networks, Hidden Markov Modeling, Support Vector Machines, etc. 4. Functional Annotation: For novel proteins that are yet to be characterized, the potential functions can be predicted by techniques such as Homology Modelling which provide an initial insight into the protein’s properties.
1 2 3 4 5 Definitions of the components Part 2 – Uses of structural databases 5. Gene Ontology: Also known as GO terms, they are identifiers to represent a gene’s functional properties categorized to cover three domains namely, “cellular component”, “molecular function” and “biological process”. 6. Root Mean Square Deviation (RMSD): Qauantification of the average distance between the atoms of the super-imposed proteins. The higher is the RMSD value, the lower is the similarity. 7. Protein Structural Alignment Server: Web based servers which help in determining the structural similarity of two given proteins by superimposing the two proteins and calculating various comparative parameters. Currently there a large number of web based servers assigned for this task. Few examples of available servers for this include DALI (Distance Matrix Alignment), MAMMOTH (Matching Molecular Models Obtained from Theory), CE/CE-MC (Combinatorial Extension -- Monte Carlo), SSAP(Sequential Structure Alignment Program), Pro. Fit (Protein least-squares Fitting), etc.
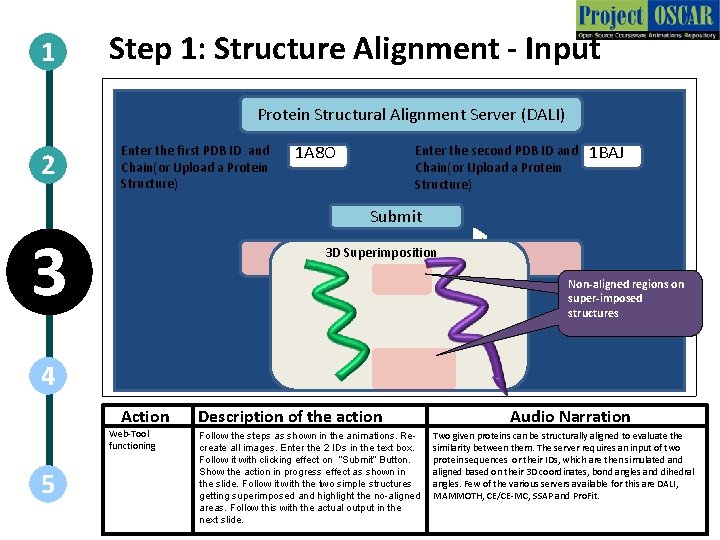
1 Step 1: Structure Alignment - Input Protein Structural Alignment Server (DALI) 2 Enter the first PDB ID and Chain(or Upload a Protein Structure) 1 A 8 O Enter the second PDB ID and Chain(or Upload a Protein Structure) 1 BAJ Submit 3 3 D Superimposition Running the Server… Non-aligned regions on super-imposed structures 4 Action Web-Tool functioning 5 Description of the action Follow the steps as shown in the animations. Recreate all images. Enter the 2 IDs in the text box. Follow it with clicking effect on “Submit” Button. Show the action in progress effect as shown in the slide. Follow it with the two simple structures getting superimposed and highlight the no-aligned areas. Follow this with the actual output in the next slide. Audio Narration Two given proteins can be structurally aligned to evaluate the similarity between them. The server requires an input of two protein sequences or their IDs, which are then simulated and aligned based on their 3 D coordinates, bond angles and dihedral angles. Few of the various servers available for this are DALI, MAMMOTH, CE/CE-MC, SSAP and Pro. Fit.
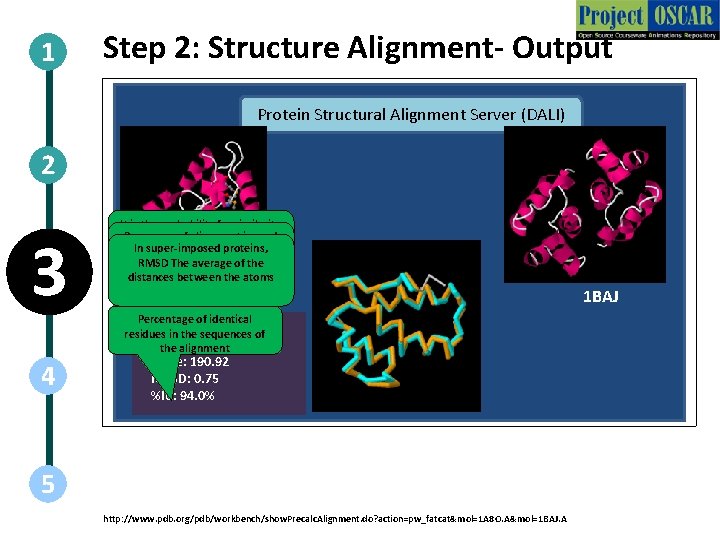
1 Step 2: Structure Alignment- Output Protein Structural Alignment Server (DALI) 2 3 4 It is the probability for similarity Raw scorethe of two alignment is used between structures. PIn super-imposed proteins, to compare other similarity value < 0. 05 indicates significant RMSD The average of the matches with same proteins similarity distances between the atoms 1 A 8 O Percentage of identical residues in the sequences of P-value: 0. 00 e+00 the alignment Score: 190. 92 RMSD: 0. 75 %Id: 94. 0% 5 http: //www. pdb. org/pdb/workbench/show. Precalc. Alignment. do? action=pw_fatcat&mol=1 A 8 O. A&mol=1 BAJ. A 1 BAJ
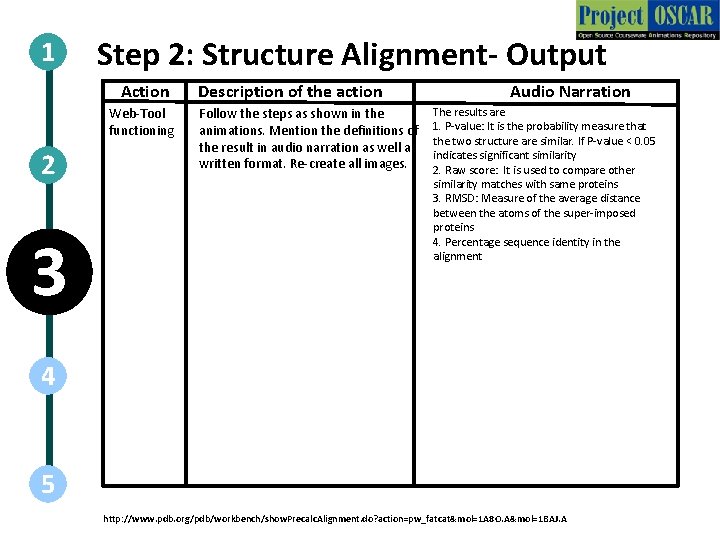
1 Step 2: Structure Alignment- Output Action Web-Tool functioning 2 3 Description of the action Follow the steps as shown in the animations. Mention the definitions of the result in audio narration as well as written format. Re-create all images. Audio Narration The results are 1. P-value: It is the probability measure that the two structure are similar. If P-value < 0. 05 indicates significant similarity 2. Raw score: It is used to compare other similarity matches with same proteins 3. RMSD: Measure of the average distance between the atoms of the super-imposed proteins 4. Percentage sequence identity in the alignment 4 5 http: //www. pdb. org/pdb/workbench/show. Precalc. Alignment. do? action=pw_fatcat&mol=1 A 8 O. A&mol=1 BAJ. A
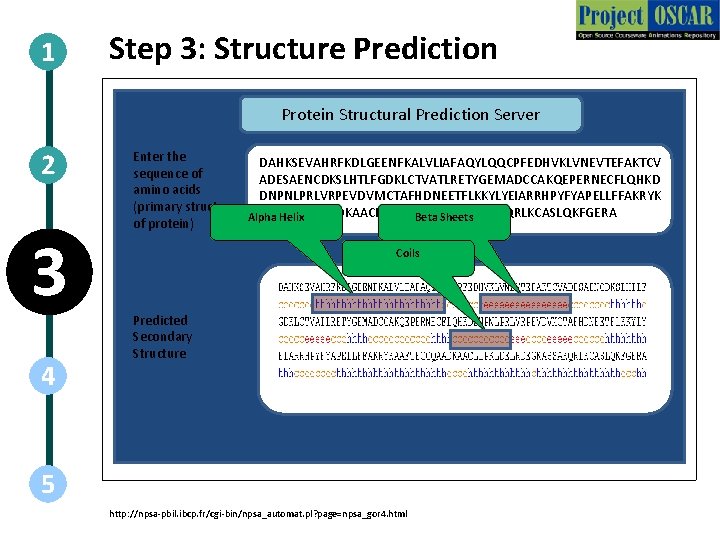
1 Step 3: Structure Prediction Protein Structural Prediction Server 2 3 4 Enter the DAHKSEVAHRFKDLGEENFKALVLIAFAQYLQQCPFEDHVKLVNEVTEFAKTCV sequence of ADESAENCDKSLHTLFGDKLCTVATLRETYGEMADCCAKQEPERNECFLQHKD amino acids DNPNLPRLVRPEVDVMCTAFHDNEETFLKKYLYEIARRHPYFYAPELLFFAKRYK (primary structure AAFTECCQAADKAACLLPKLDELRDEGKASSAKQRLKCASLQKFGERA Alpha Helix Beta Sheets of protein) Coils Predicted Secondary Structure 5 http: //npsa-pbil. ibcp. fr/cgi-bin/npsa_automat. pl? page=npsa_gor 4. html
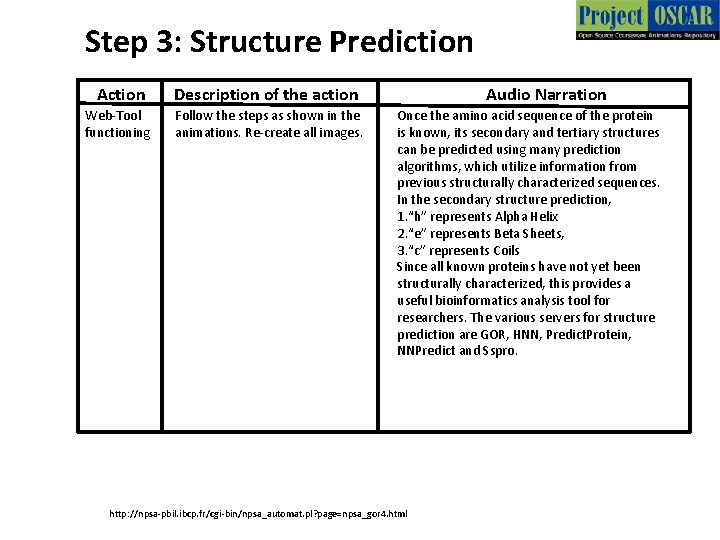
Step 3: Structure Prediction Action Description of the action Web-Tool functioning Follow the steps as shown in the animations. Re-create all images. Audio Narration Once the amino acid sequence of the protein is known, its secondary and tertiary structures can be predicted using many prediction algorithms, which utilize information from previous structurally characterized sequences. In the secondary structure prediction, 1. “h” represents Alpha Helix 2. “e” represents Beta Sheets, 3. “c” represents Coils Since all known proteins have not yet been structurally characterized, this provides a useful bioinformatics analysis tool for researchers. The various servers for structure prediction are GOR, HNN, Predict. Protein, NNPredict and Sspro. http: //npsa-pbil. ibcp. fr/cgi-bin/npsa_automat. pl? page=npsa_gor 4. html
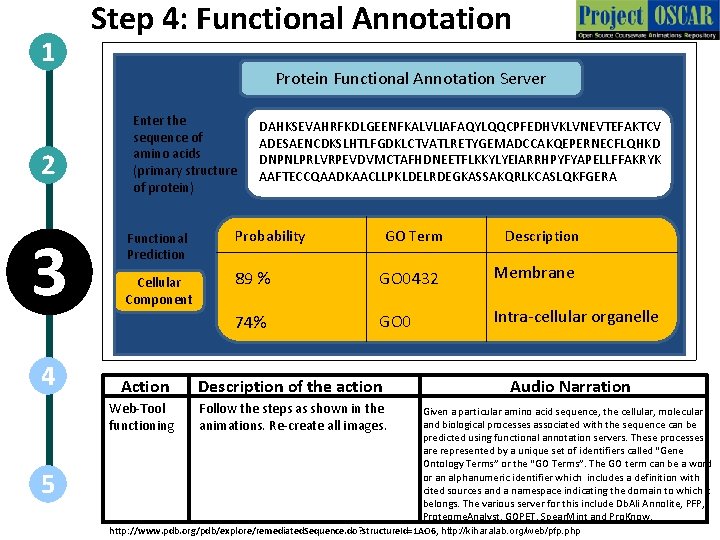
1 2 3 4 5 Step 4: Functional Annotation Protein Functional Annotation Server Enter the sequence of amino acids (primary structure of protein) DAHKSEVAHRFKDLGEENFKALVLIAFAQYLQQCPFEDHVKLVNEVTEFAKTCV ADESAENCDKSLHTLFGDKLCTVATLRETYGEMADCCAKQEPERNECFLQHKD DNPNLPRLVRPEVDVMCTAFHDNEETFLKKYLYEIARRHPYFYAPELLFFAKRYK AAFTECCQAADKAACLLPKLDELRDEGKASSAKQRLKCASLQKFGERA Functional Prediction Probability GOTerm GO Molecular Biological Cellular Component Functions 100 89 100 %%% GO 0189 GO 0432 GO 0549 C 21 Steroid Hormone Vitamin Membrane D Binding 97% 74% 97% GO 0243 GO 0543 Water Bindingorganelle Intra-cellular Action Description of the action Web-Tool functioning Follow the steps as shown in the animations. Re-create all images. Description Metabolism Vitamin Transport Audio Narration Given a particular amino acid sequence, the cellular, molecular and biological processes associated with the sequence can be predicted using functional annotation servers. These processes are represented by a unique set of identifiers called “Gene Ontology Terms” or the “GO Terms”. The GO term can be a word or an alphanumeric identifier which includes a definition with cited sources and a namespace indicating the domain to which it belongs. The various server for this include Db. Ali Annolite, PFP, Proteome. Analyst, GOPET, Spear. Mint and Pro. Know. http: //www. pdb. org/pdb/explore/remediated. Sequence. do? structure. Id=1 AO 6, http: //kiharalab. org/web/pfp. php
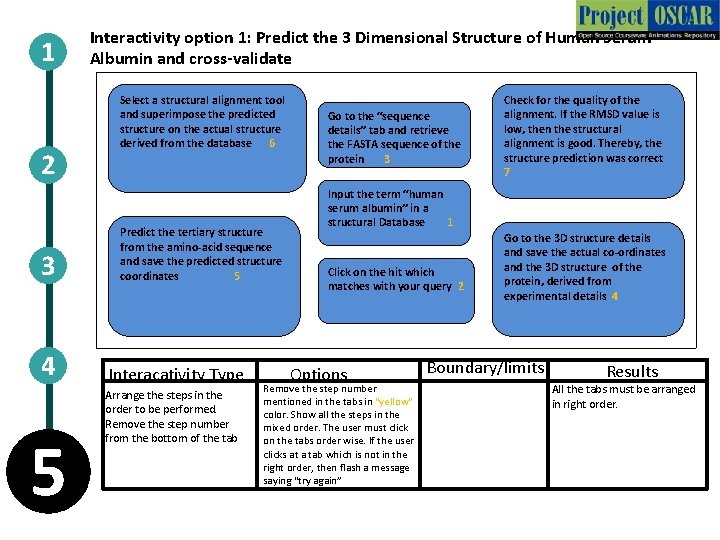
1 2 3 4 5 Interactivity option 1: Predict the 3 Dimensional Structure of Human Serum Albumin and cross-validate Select a structural alignment tool and superimpose the predicted structure on the actual structure derived from the database 6 Predict the tertiary structure from the amino-acid sequence and save the predicted structure coordinates 5 Interacativity Type Arrange the steps in the order to be performed. Remove the step number from the bottom of the tab Go to the “sequence details” tab and retrieve the FASTA sequence of the protein 3 Check for the quality of the alignment. If the RMSD value is low, then the structural alignment is good. Thereby, the structure prediction was correct 7 Input the term “human serum albumin” in a structural Database 1 Click on the hit which matches with your query 2 Options Remove the step number mentioned in the tabs in “yellow” color. Show all the steps in the mixed order. The user must click on the tabs order wise. If the user clicks at a tab which is not in the right order, then flash a message saying “try again” Go to the 3 D structure details and save the actual co-ordinates and the 3 D structure of the protein, derived from experimental details 4 Boundary/limits Results All the tabs must be arranged in right order.
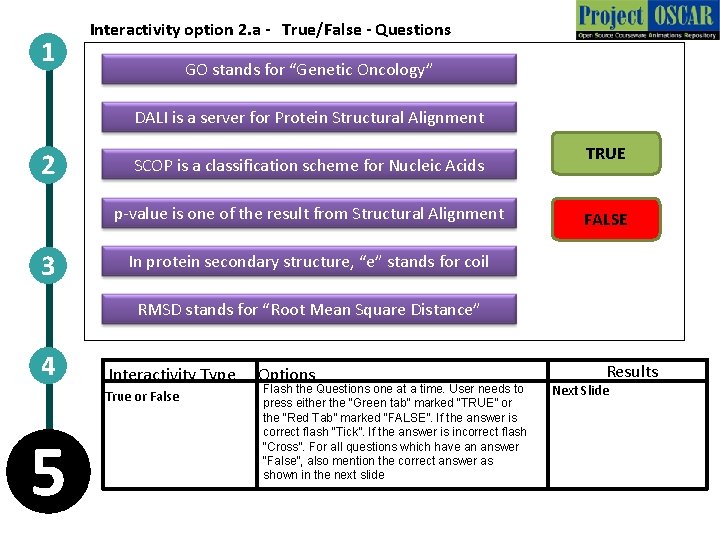
1 Interactivity option 2. a - True/False - Questions GO stands for “Genetic Oncology” DALI is a server for Protein Structural Alignment 2 SCOP is a classification scheme for Nucleic Acids p-value is one of the result from Structural Alignment 3 TRUE FALSE In protein secondary structure, “e” stands for coil RMSD stands for “Root Mean Square Distance” 4 Interactivity Type True or False 5 Options Flash the Questions one at a time. User needs to press either the “Green tab” marked “TRUE” or the “Red Tab” marked “FALSE”. If the answer is correct flash “Tick”. If the answer is incorrect flash “Cross”. For all questions which have an answer “False”, also mention the correct answer as shown in the next slide Results Next Slide
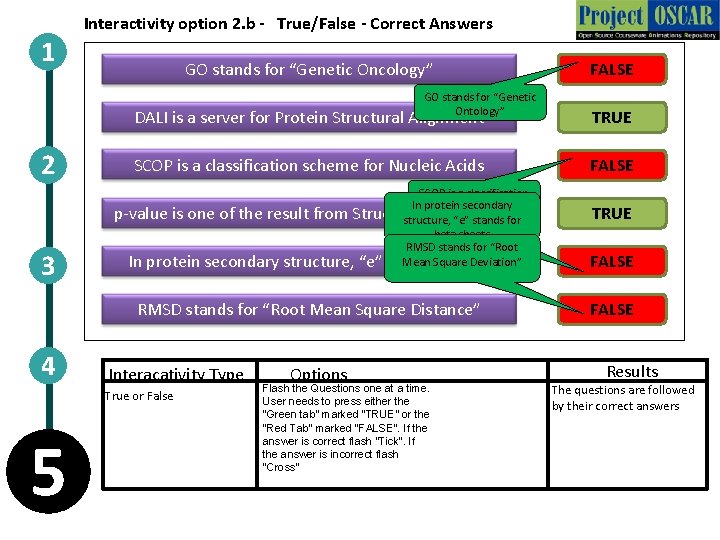
1 Interactivity option 2. b - True/False - Correct Answers GO stands for “Genetic Oncology” GO stands for “Genetic Ontology” 2 3 DALI is a server for Protein Structural Alignment TRUE SCOP is a classification scheme for Nucleic Acids FALSE SCOP is a classification In protein schemesecondary for Proteins p-value is one of the result from Structural Alignment structure, “e” stands for beta sheets RMSD stands for “Root Mean Square Deviation” In protein secondary structure, “e” stands for coil RMSD stands for “Root Mean Square Distance” 4 Interacativity Type True or False 5 FALSE Options Flash the Questions one at a time. User needs to press either the “Green tab” marked “TRUE” or the “Red Tab” marked “FALSE”. If the answer is correct flash “Tick”. If the answer is incorrect flash “Cross” TRUE FALSE Results The questions are followed by their correct answers
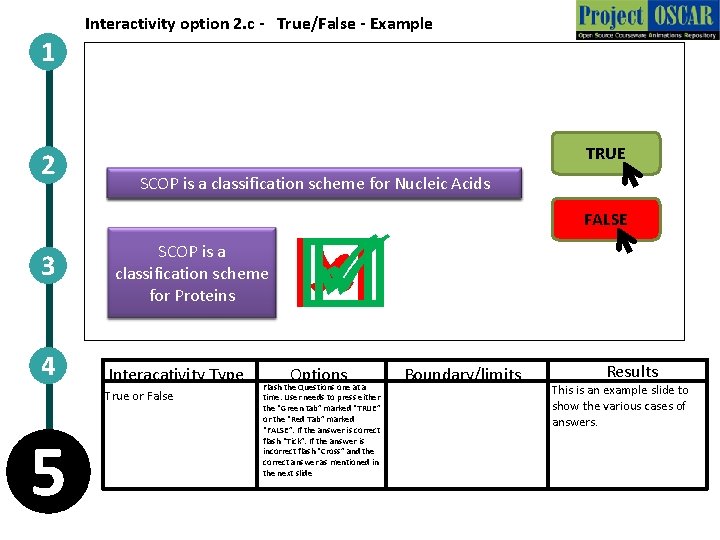
1 2 3 4 Interactivity option 2. c - True/False - Example TRUE SCOP DALI isis. GO aaserver classification stands forfor Protein “Genetic scheme Structural Oncology” for Nucleic Alignment Acids The correct answer SCOP is a is “False”. GO stands classification scheme for “Genetic for Proteins Ontology” Interacativity Type True or False 5 FALSE Options Flash the Questions one at a time. User needs to press either the “Green tab” marked “TRUE” or the “Red Tab” marked “FALSE”. If the answer is correct flash “Tick”. If the answer is incorrect flash “Cross” and the correct answer as mentioned in the next slide Boundary/limits Results This is an example slide to show the various cases of answers.

1 Questionnaire 1. Which is the server for Protein Structure Prediction ? 2 Answers: a) Prot. Param d) DALI 4 c) nn. PREDICT 2. Which is the server for Functional annotation of Proteins? Answers: a) DALI Analyst 3 b) Peptide. Mass b) GOR c) SSAP d) Proteome 3. Which amongst these is NOT the output for Functional annotation? Answers: a) GO Term b)Source Organism of annotation d) Description of Function 4. By default, PDB structures appear in which visualization tool? Answers: a) VMD above b) NAMD c) Jmol d) None of the 5. PDB is primarily which Database? a) Protein b) Nucleotide c) Gene d) None of the Above 5 c) Probability

Links for further reading Reference websites http: //npsa-pbil. ibcp. fr/cgi-bin/npsa_automat. pl? page=npsa_gor 4. html http: //cubic. bioc. columbia. edu/predictprotein/ http: //ekhidna. biocenter. helsinki. fi/dali_lite/start http: //kiharalab. org/web/pfp. php http: //pa. cs. ualberta. ca: 8080/pa/index. html http: //www. ebi. ac. uk/Tools/clustalw 2/index. html http: //www. pdb. org/pdb/home. do http: //expasy. org/sprot/ http: //expasy. org/prosite/ http: //webdocs. ualberta. ca/~bioinfo/PA/

Links for further reading Following URLs are used for animations http: //www. pdb. org/pdb/search/adv. Search. do http: //www. pdb. org/pdb/explore. do? structure. Id=1 AUM http: //www. pdb. org/pdb/workbench/show. Precalc. Alignment. do? action =pw_fatcat&mol=1 A 8 O. A&mol=1 BAJ. A http: //npsa-pbil. ibcp. fr/cgi-bin/npsa_automat. pl? page=npsa_gor 4. html http: //www. pdb. org/pdb/explore/remediated. Sequence. do? structure. Id =1 AO 6 http: //kiharalab. org/web/pfp. php

Links for further reading Published Literature SCOP: A Structural Classification of Proteins Database for the Investigation of Sequences and Structures Alexey G. Murzin, Steven E. Brenner, Tim Hubbard and Cyrus Chothia. J. Mol. Biol. (1995) 247, 536– 540 CATH — a hierarchic classification of protein domain structures CA Orengo, AD Michie, S Jones, DT Jones, MB Swindells and JM Thornton Structure 1997, Vol 5 No 8 Books: Bioinformatics Sequence and Genome Analysis by David Mount
- Slides: 32