Bioinformatics analysis pipeline for viral metagenomics Davit Bzhalava
Bioinformatics analysis pipeline for viral metagenomics Davit Bzhalava, Ph. D Dept. of Laboratory Medicine, Karolinska Institutet, Sweden Davit Bzhalava 9/24/2021 1

Human Microbiota Ø We are born 100% human and we die 90% microbial. Ø The term human microbiome or microbiota, defines the collection of microorganisms that reside in the human body. Ø The viral fraction of human microbiome is referred to as the human virome. Ø Viruses constitute only a small part of human microbiota, but their proportion and composition seems to change in diseased individuals. Davit Bzhalava 9/24/2021 2
Tumor Viruses Ø 2 million (16%) of new cancer cases worldwide was estimated to be attributable to infections in 2008. Ø 1300000 (65%) of these cancers were attributable to viral infections Ø There is epidemiological indication that additional cancerassociated viruses may exist: § Increased incidence of some cancer types among immunosuppressed individuals; § Space and time clustering of childhood leukemias. Davit Bzhalava 9/24/2021 3

Purpose of viral metagenomics Ø Who is there? Ø What are they doing? Ø How are they doing it? Davit Bzhalava 9/24/2021 4
Needle in a haystack Ø Viruses usually constitute <0. 1% of the whole metagenomic datasets Ø Small changes in the data analysis pipeline can drastically alter results Davit Bzhalava 9/24/2021 5

Bioinformatics Pipeline Library Preparation Filter out Human, bacterial, phage and vector sequences Assembly validation & number of reads estimation Data Analysis Sequencing Normalize k-mer frequencies Genome assembly Taxonomic classification Final characterization of virus related sequences Case-control comparison of virus related & “unknown” sequences / OR estimation Davit Bzhalava 9/24/2021 6

Bioinformatics Pipeline Library Preparation Filter out Human, bacterial, phage and vector sequences Assembly validation & number of reads estimation Sequencing Data Analysis Normalize kmer frequencies Genome assembly Taxonomic classification Final characterization of virus related sequences Case-control comparison of virus related & “unknown” sequences / OR estimation Davit Bzhalava 9/24/2021 7

de novo assembly NGS technologies produce billions of short reads from random locations in the genome by oversampling it. Assembly algorithms, in the process called de novo assembly, reconstruct original genomes present in the sample by merging short genomic fragments into longer contiguous sequences (“contigs”). There are two main types of de novo assembly programs: ü Overlap/Layout/Consensus (OLC) assemblers ü de Bruijn Graph Assemblers Davit Bzhalava 24/09/2021 8

OLC assembly Overlap: find potentially overlapping reads Layout: merge reads into contigs and contigs into supercontigs Consensus: derive the DNA sequence and correct read errors Davit Bzhalava . . ACGATTACAATAGGTT. .
de Bruijn graph assembly ü de Bruijn graph assemblers model the relationship between exact substrings of length k extracted from the input reads. ü In de Bruijn graph the reads themselves are not directly modelled but they are implicitly represented as paths through the de Bruijn graph. ü Most de Bruijn graph assemblers use the read information to refine the graph structure and to remove graph patterns that are not consistent with the reads. ü de Bruijn graph approach is based on exact matches, thus error correction approaches (used both before and during assembly) are crucial for achieving high-quality assemblies. Davit Bzhalava 24/09/2021 10

Challenges in assembly Ø If we have 2 sequences 1. 2. the_quick_brown_fox_jumps_over_the_lazy_dog Ø Will be decomposed into k-mers § Kmer = 5 Ø put both sentences into the same graph and follow the links in the graph § the_q -> he_qu -> e_qui -> _quic -> quick -> uick_ -> ick_b -> ck_br Ø to spell out the 'assembled' sentence, § the_quick_brown_fox_jumps_over_the_lazy_dog Ø If kmer = 6: there's no 6 -mer word that is in common between the sentence fragments. Ø If k-mer = 4, the graph becomes complicated: the word the_ appears twice ***Example taken from: http: //ivory. idyll. org/blog/the-k-parameter. html Davit Bzhalava 9/24/2021 11
Challenges in assembly Ø Solution is to try as many assemblers and with as many parameters as possible. Ø Resources including time is limited Ø Assemblies are RAM thirsty § Next. Seq, 300 m reads ≈250 GB RAM Ø kmer based assemblers scale poorly Davit Bzhalava 9/24/2021 12
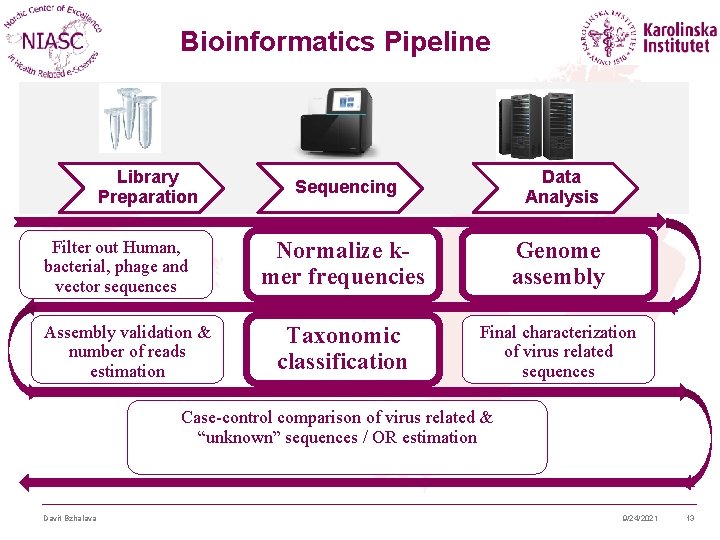
Bioinformatics Pipeline Library Preparation Filter out Human, bacterial, phage and vector sequences Assembly validation & number of reads estimation Sequencing Data Analysis Normalize kmer frequencies Genome assembly Taxonomic classification Final characterization of virus related sequences Case-control comparison of virus related & “unknown” sequences / OR estimation Davit Bzhalava 9/24/2021 13
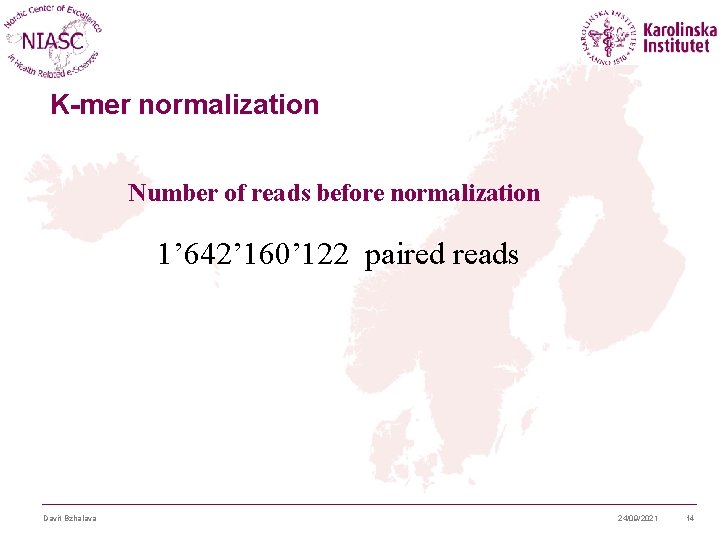
K-mer normalization Number of reads before normalization 1’ 642’ 160’ 122 paired reads Davit Bzhalava 24/09/2021 14
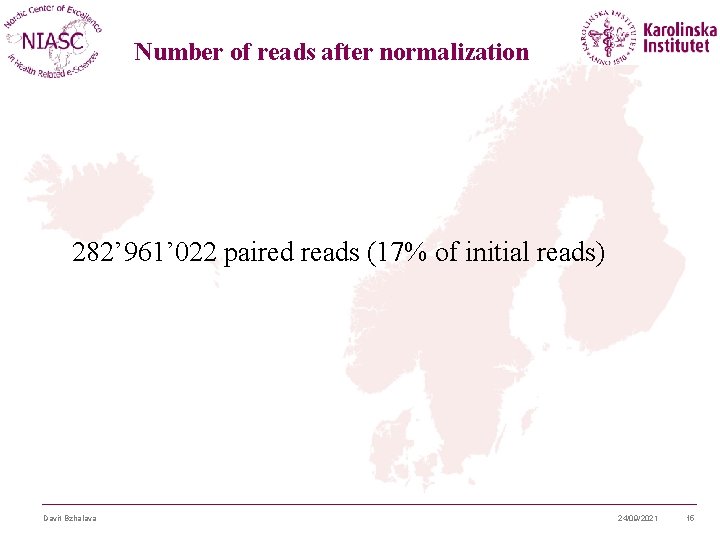
Number of reads after normalization 282’ 961’ 022 paired reads (17% of initial reads) Davit Bzhalava 24/09/2021 15

Human genome coverage before normalization Davit Bzhalava 24/09/2021 16

Human genome coverage after normalization Davit Bzhalava 24/09/2021 17
Number of reads after HG clean up 6’ 745’ 443 paired reads (0. 02 % normalized data and 0. 004% of initial reads) Davit Bzhalava 24/09/2021 18
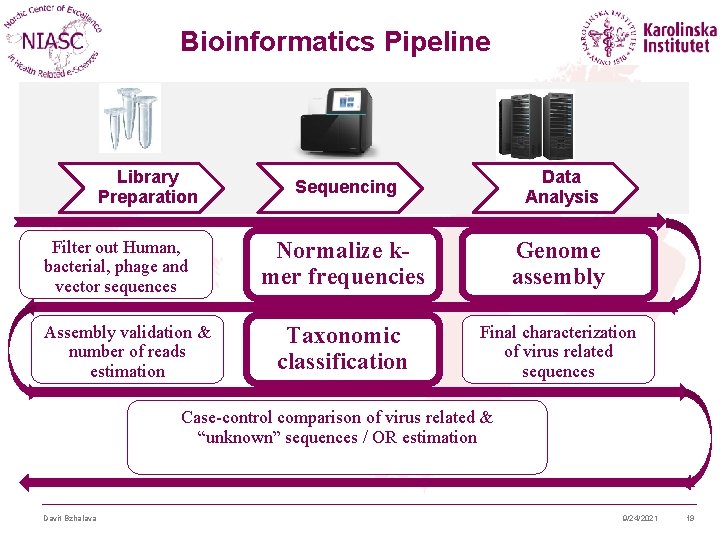
Bioinformatics Pipeline Library Preparation Filter out Human, bacterial, phage and vector sequences Assembly validation & number of reads estimation Sequencing Data Analysis Normalize kmer frequencies Genome assembly Taxonomic classification Final characterization of virus related sequences Case-control comparison of virus related & “unknown” sequences / OR estimation Davit Bzhalava 9/24/2021 19
Taxonomic classification Ø NCBI BLAST - One of the most famous similarity-based taxonomic classification Ø NCBI BLAST compares sequences to known genomes Davit Bzhalava 9/24/2021 20
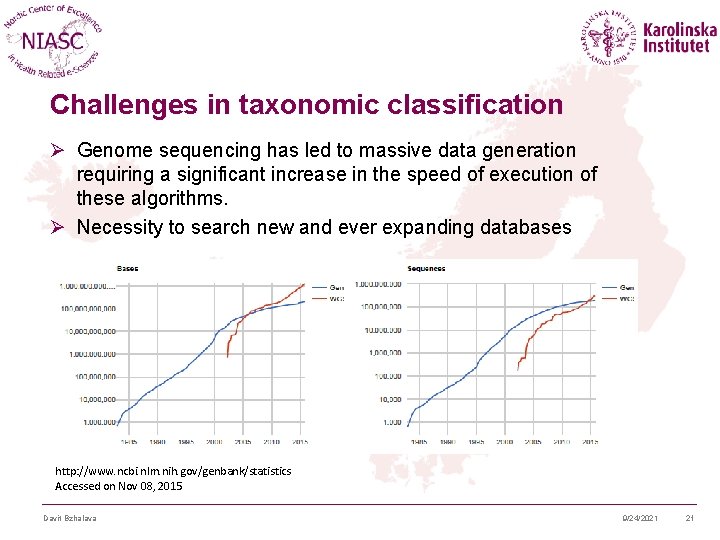
Challenges in taxonomic classification Ø Genome sequencing has led to massive data generation requiring a significant increase in the speed of execution of these algorithms. Ø Necessity to search new and ever expanding databases http: //www. ncbi. nlm. nih. gov/genbank/statistics Accessed on Nov 08, 2015 Davit Bzhalava 9/24/2021 21

Challenges in taxonomic classification Ø NCBI BLAST-based search tools § are extremely time consuming § may take days or even weeks to complete when large metagenomic datasets need to be compared against nucleotide or protein databases Ø Paracel Blast a commercial software § Achieved the same results, on same file, on same machine 10 times faster Ø Scalable open source NCBI BLAST solutions are needed Davit Bzhalava 9/24/2021 22

Thank you! Davit Bzhalava 9/24/2021 23
- Slides: 23