Biodegradation of 1 4 Dioxane A Microbiologists Perspective
Biodegradation of 1, 4 -Dioxane A Microbiologist’s Perspective Mike Hyman Department of Plant and Microbial Biology NC State University mrhyman@ncsu. edu
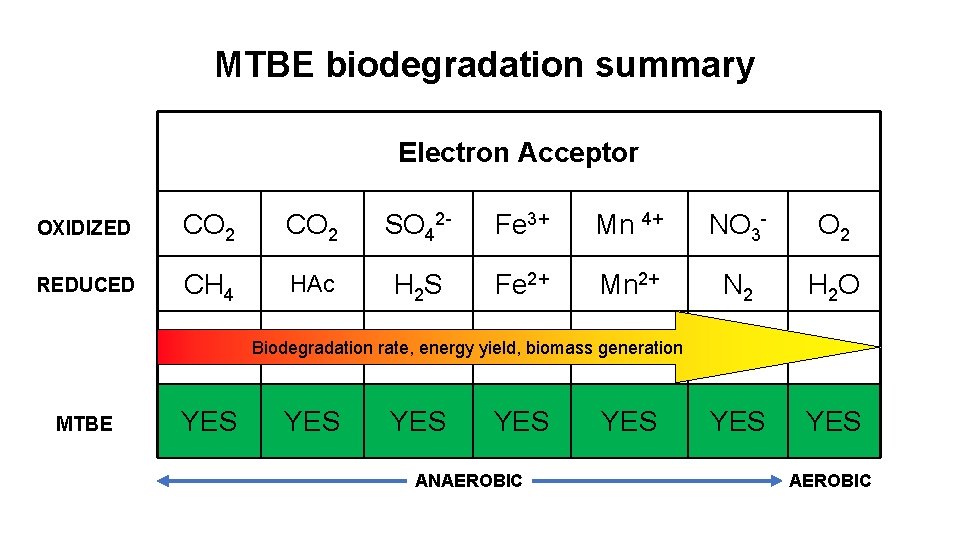
MTBE biodegradation summary Electron Acceptor OXIDIZED CO 2 SO 42 - Fe 3+ Mn 4+ NO 3 - O 2 REDUCED CH 4 HAc H 2 S Fe 2+ Mn 2+ N 2 H 2 O YES Biodegradation rate, energy yield, biomass generation MTBE YES YES ANAEROBIC YES AEROBIC
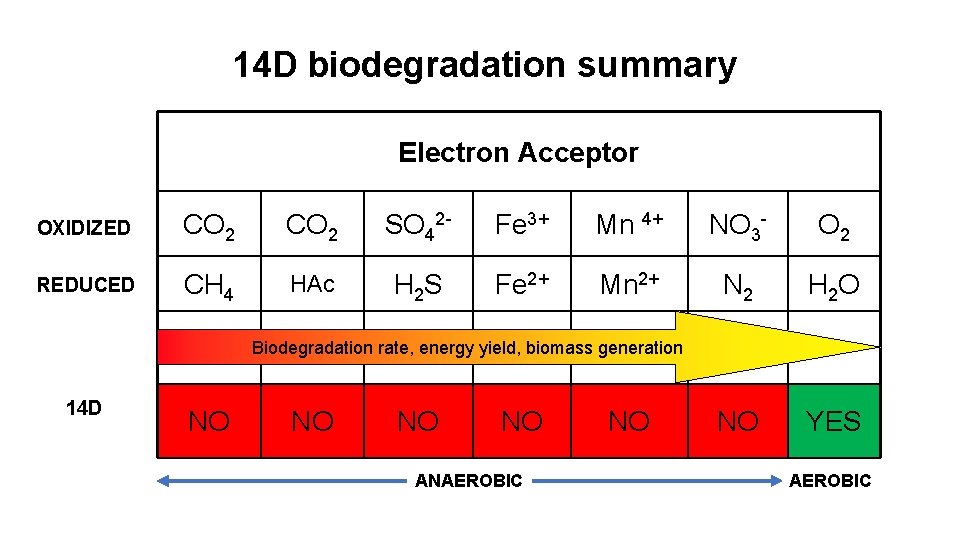
14 D biodegradation summary Electron Acceptor OXIDIZED CO 2 SO 42 - Fe 3+ Mn 4+ NO 3 - O 2 REDUCED CH 4 HAc H 2 S Fe 2+ Mn 2+ N 2 H 2 O NO YES Biodegradation rate, energy yield, biomass generation 14 D NO NO ANAEROBIC NO AEROBIC

Aerobic Metabolism: A conventional model for biodegradation • Microorganisms use Co. C as source of carbon and energy for growth • • Microbial growth is directly related to decrease in Co. C concentration Major metabolite release (other than CO 2) is typically limited Mineralization to CO 2 requires whole pathways Bacteria may have “evolved” to metabolize Co. C Bacteria that metabolize Co. Cs may be quite rare Bacteria have maintenance energy requirements that must be met If Co. C is a poor substrate, Smin and Ks will likely be high
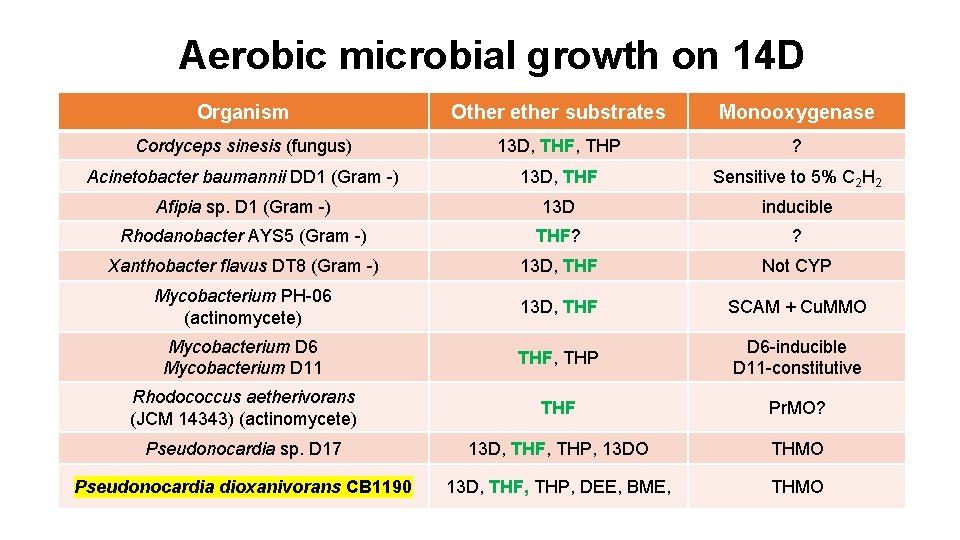
Aerobic microbial growth on 14 D Organism Other ether substrates Monooxygenase Cordyceps sinesis (fungus) 13 D, THF, THP ? Acinetobacter baumannii DD 1 (Gram -) 13 D, THF Sensitive to 5% C 2 H 2 Afipia sp. D 1 (Gram -) 13 D inducible Rhodanobacter AYS 5 (Gram -) THF? ? Xanthobacter flavus DT 8 (Gram -) 13 D, THF Not CYP Mycobacterium PH-06 (actinomycete) 13 D, THF SCAM + Cu. MMO Mycobacterium D 6 Mycobacterium D 11 THF, THP D 6 -inducible D 11 -constitutive Rhodococcus aetherivorans (JCM 14343) (actinomycete) THF Pr. MO? Pseudonocardia sp. D 17 13 D, THF, THP, 13 DO THMO Pseudonocardia dioxanivorans CB 1190 13 D, THF, THP, DEE, BME, THMO
A cross country THF-metabolizer gone rogue! Pseudonocardia dioxanivorans CB 1190 (Mahendra & Alvarez–Cohen, 2005) (CA) Amycolata sp. CB 1190 (Kelley et al. , 2001) (IA) Strain CB 1190 (ATCC 55486) Pseudonocardia (Amycolata) (Parales et al. , 1994) (SC) Mixed culture growing on THF
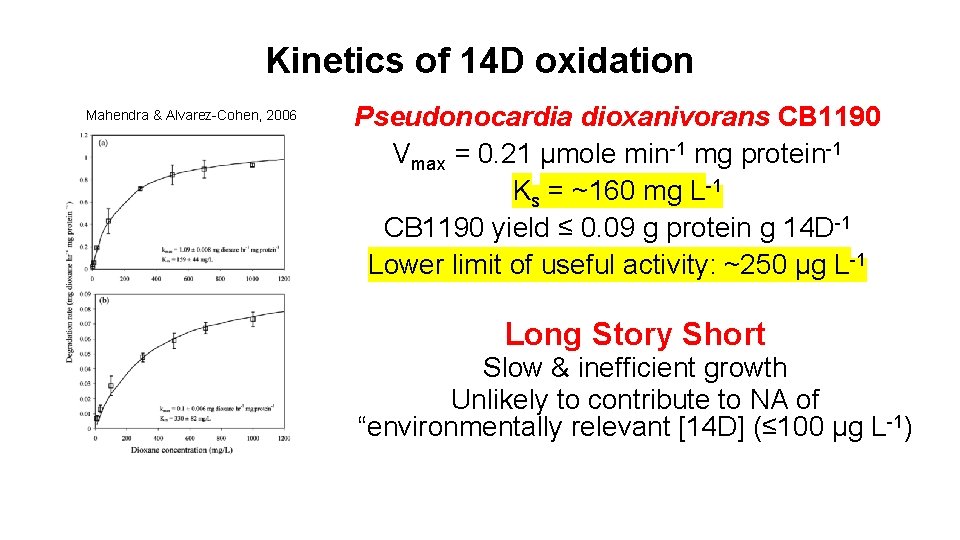
Kinetics of 14 D oxidation Mahendra & Alvarez-Cohen, 2006 Pseudonocardia dioxanivorans CB 1190 Vmax = 0. 21 µmole min-1 mg protein-1 Ks = ~160 mg L-1 CB 1190 yield ≤ 0. 09 g protein g 14 D-1 Lower limit of useful activity: ~250 µg L-1 Long Story Short Slow & inefficient growth Unlikely to contribute to NA of “environmentally relevant [14 D] (≤ 100 µg L-1)

Aerobic Cometabolism: A less conventional biodegradation model • • Microorganisms do not use Co. C as growth-supporting compound Microbial growth is supported by 1 o substrate (e. g. methane, propane) Major metabolite release (other than CO 2) is common (in pure culture) Activity relies on small number of enzymes (1 o substrate induction) No “evolution” needed (but key enzymes often plasmid-encoded) Bacteria that cometabolize Co. Cs can be common/ubiquitous Maintenance energy requirements are supported by 1 o substrate Capable of degrading Co. Cs to very low concentrations (ppb-ppt)
![1 o Substrate summary with pure cultures High [14 D] >100 ppm Low [14 1 o Substrate summary with pure cultures High [14 D] >100 ppm Low [14](http://slidetodoc.com/presentation_image_h2/2777a6b308c07353c4583f0dc3a28787/image-9.jpg)
1 o Substrate summary with pure cultures High [14 D] >100 ppm Low [14 D] <100 ppb ethene, propene Sort of, maybe NO methane Sort of, maybe NO ethane YES propane YES n-butane YES isobutane YES toluene YES NO phenol NO NO tetrahydrofuran YES ammonia NO NO 1 o Substrate Depends on which bacterium
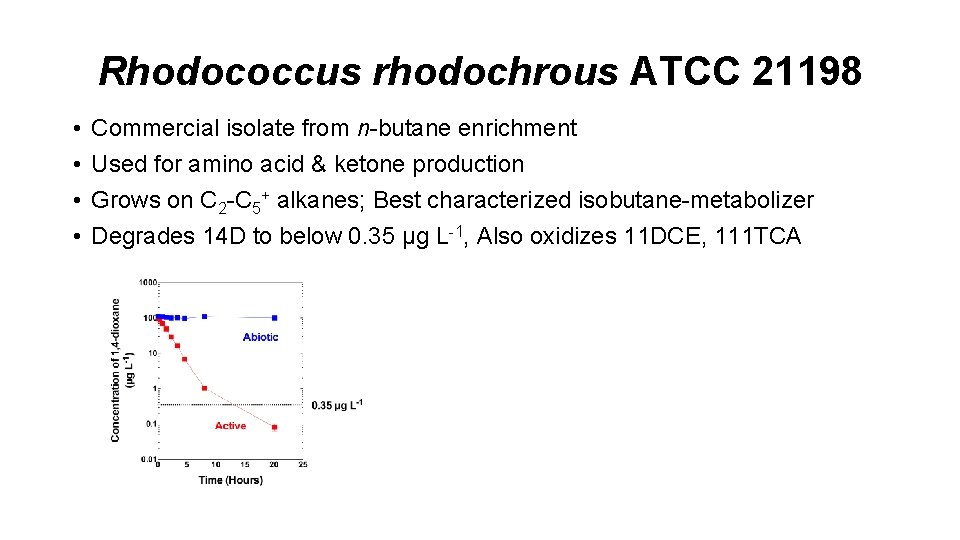
Rhodococcus rhodochrous ATCC 21198 • • Commercial isolate from n-butane enrichment Used for amino acid & ketone production Grows on C 2 -C 5+ alkanes; Best characterized isobutane-metabolizer Degrades 14 D to below 0. 35 µg L-1, Also oxidizes 11 DCE, 111 TCA
Aerobic cometabolism summary • Fortuitous biotransformation of target pollutant(s) • Separates carbon and energy acquisition from biotransformation • Potential to treat very low contaminant concentrations (ppb to ppt) • Active microorganisms are often widely distributed • Monooxygenases are common catalysts in aerobic systems • Active bacteria can often concurrently degrade multiple contaminants
EMDs for 14 D Biodegradation • Compound Specific Isotope Analysis (CSIA): • Determines effects of biodegradation on Co. C isotopic composition • Recently described for “metabolism” and “cometabolism” • Broad range of undegraded isotope signatures • Quantitative polymerase chain reaction (q. PCR): • Industry standard approach, amplifies/quantifies specific genes • Sensitive, commercially available, looks for what is already known • Confusing nomenclature (THFMO = DXMO, Pr. MO ≠ Pr. MO) • Emerging EMDs • Proteomic approaches (actual vs potential expression) • 14 C-based assays (slow rates of degradation)
- Slides: 12