Bio SEAL InMemory Biological Sequence Alignment Accelerator for
Bio. SEAL: In-Memory Biological Sequence Alignment Accelerator for Large-Scale Genomic Data Roman Kaplan, Leonid Yavits and Ran Ginosar Department of Electrical Engineering Technion

Part 1: Background and Previous Work 1. Motivation 2. Background: a) Content Addressable Memory (CAM) b) Associative Processing c) Memristors 3. Previous work: Resistive CAM (Re. CAM)
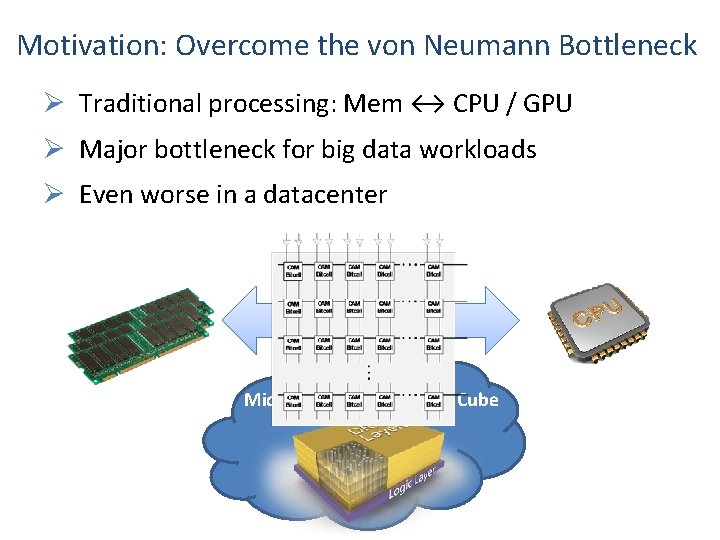
Motivation: Overcome the von Neumann Bottleneck Ø Traditional processing: Mem ↔ CPU / GPU Ø Major bottleneck for big data workloads Ø Even worse in a datacenter 11000101… Micron’s Hybrid Memory Cube
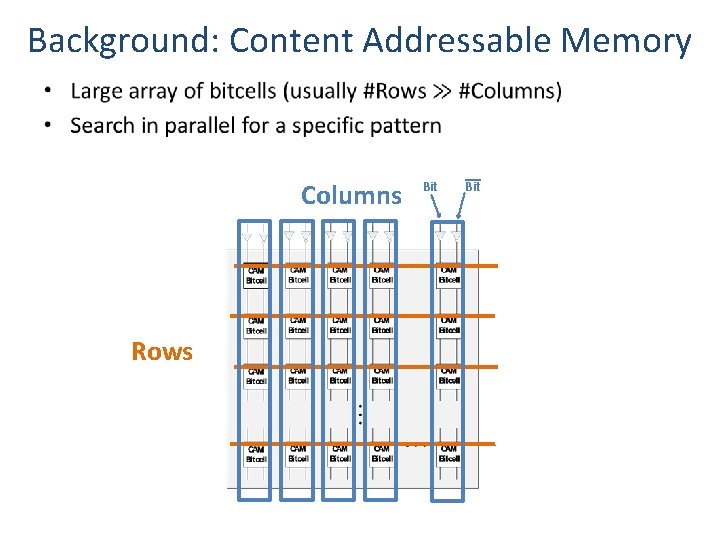
Background: Content Addressable Memory • Columns Rows Bit
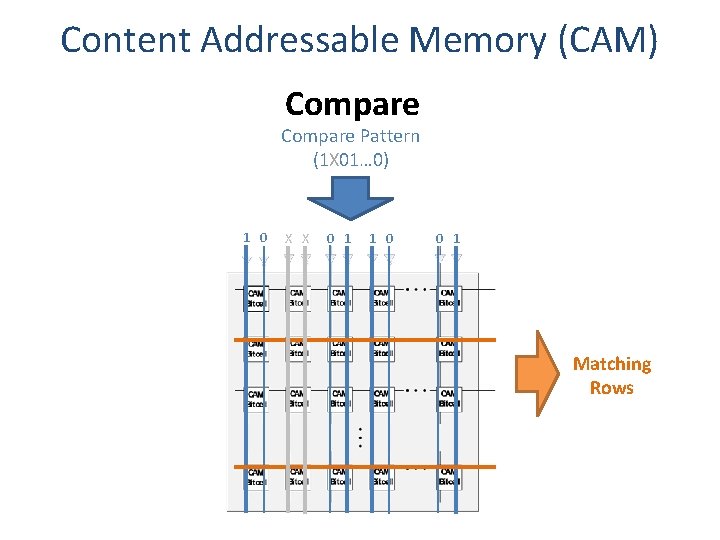
Content Addressable Memory (CAM) Compare Pattern (1 X 01… 0) 1 0 X X 0 1 1 0 0 1 Matching Rows
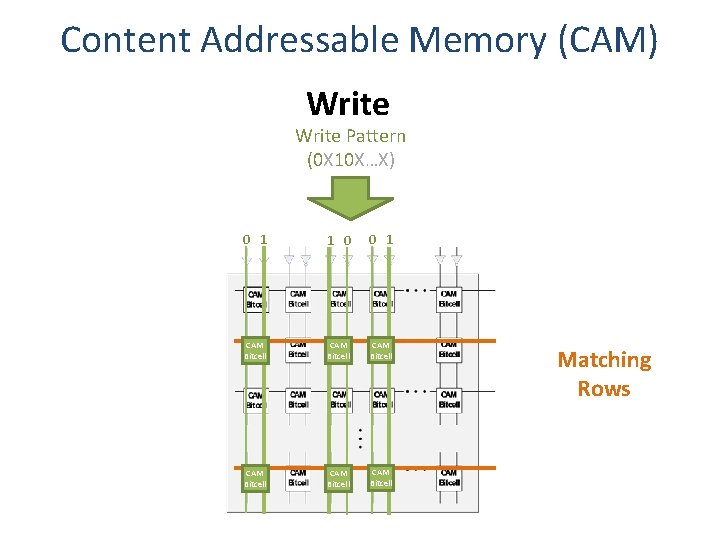
Content Addressable Memory (CAM) Write Pattern (0 X 10 X…X) 0 1 1 0 0 1 CAM Bitcell CAM Bitcell Matching Rows
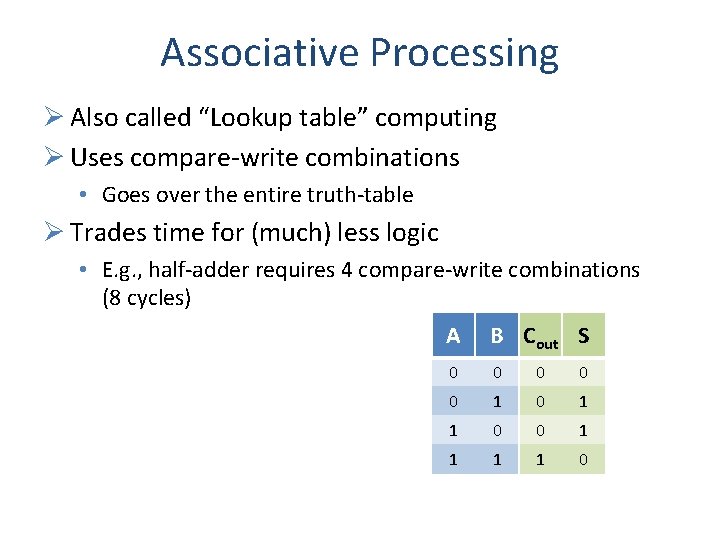
Associative Processing Ø Also called “Lookup table” computing Ø Uses compare-write combinations • Goes over the entire truth-table Ø Trades time for (much) less logic • E. g. , half-adder requires 4 compare-write combinations (8 cycles) A B Cout S 0 0 0 1 1 0
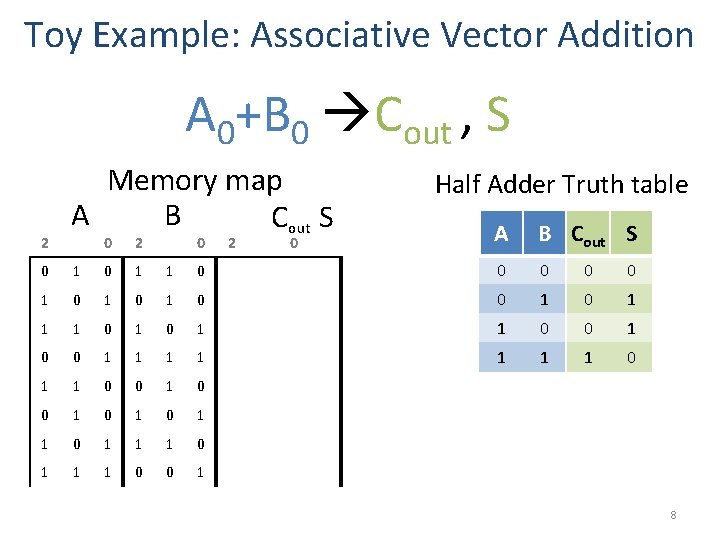
Toy Example: Associative Vector Addition A 0+B 0 Cout , S 2 Memory map A B Cout S 0 2 0 Half Adder Truth table A B Cout S 0 1 1 0 0 0 1 0 1 0 1 1 0 0 1 1 1 1 0 0 1 0 1 1 1 0 0 1 8

Example: Associative Vector Addition Compare 2 Memory map A B Cout S 0 2 0 3 Write Half Adder Truth table 0 A B Cout S 0 0 0 0 1 0 1 0 1 1 0 0 1 1 1 0 0 1 0 1 0 0 0 1 1 1 0 0 1 1 0 0 1 1 Evaluation 1 0 1 1 0 0 0 1 9
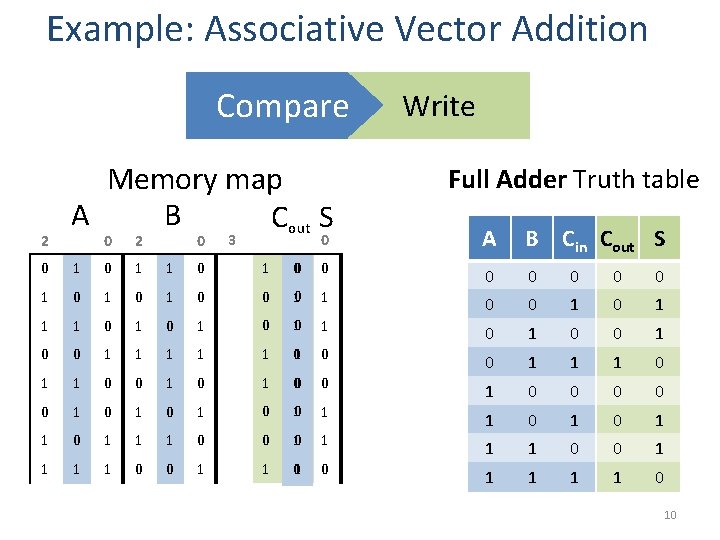
Example: Associative Vector Addition Compare 2 Memory map A B Cout S 0 2 0 3 Write Full Adder Truth table 0 A B Cin Cout S 0 1 1 0 0 0 0 1 0 10 0 1 1 0 1 0 1 10 0 1 1 1 1 0 1 0 0 1 0 0 1 1 1 0 0 0 1 0 10 1 1 0 0 1 0 1 1 1 0 0 10 1 1 0 0 1 1 01 0 1 1 0 0 1 1 1 0 10
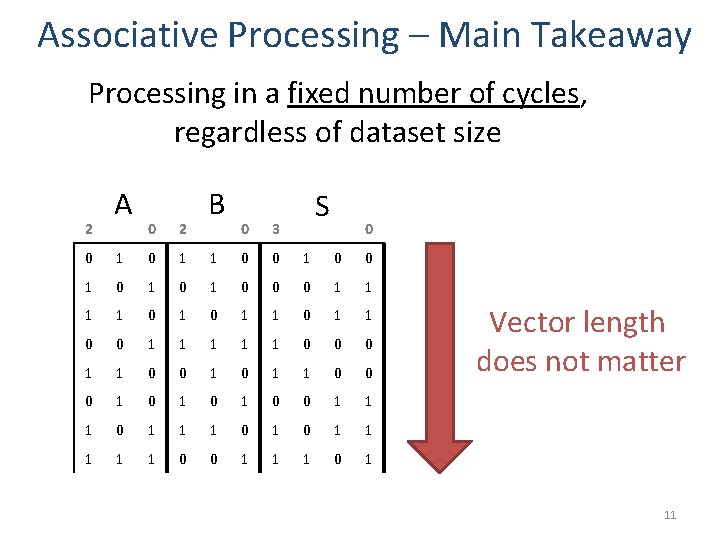
Associative Processing – Main Takeaway Processing in a fixed number of cycles, regardless of dataset size 2 A 0 2 B 0 3 S 0 0 1 1 0 0 1 0 1 0 0 0 1 1 0 1 1 0 0 1 1 1 0 0 0 1 1 0 0 0 1 0 1 0 0 1 1 1 0 0 1 1 1 0 1 Vector length does not matter 11
So What’s New? Memristors Ø Devices that change resistance with applied voltage Ø Non volatile Ø Can be placed in metal layers above silicon Ø Small area footprint: 4 F 2
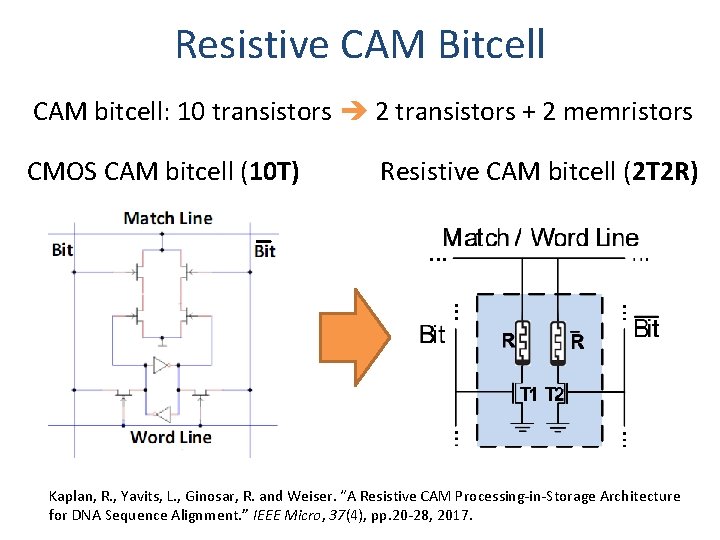
Resistive CAM Bitcell CAM bitcell: 10 transistors 2 transistors + 2 memristors CMOS CAM bitcell (10 T) Resistive CAM bitcell (2 T 2 R) Kaplan, R. , Yavits, L. , Ginosar, R. and Weiser. “A Resistive CAM Processing-in-Storage Architecture for DNA Sequence Alignment. ” IEEE Micro, 37(4), pp. 20 -28, 2017.
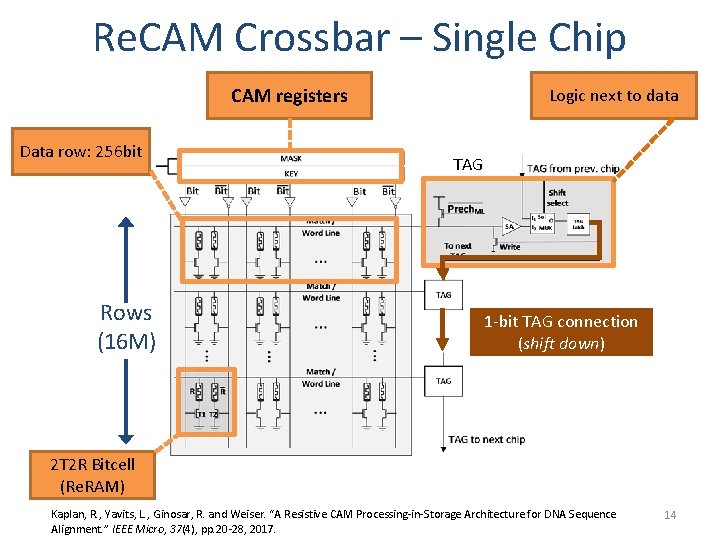
Re. CAM Crossbar – Single Chip CAM registers Data row: 256 bit Rows (16 M) Logic next to data TAG 1 -bit TAG connection (shift down) 2 T 2 R Bitcell (Re. RAM) Kaplan, R. , Yavits, L. , Ginosar, R. and Weiser. “A Resistive CAM Processing-in-Storage Architecture for DNA Sequence Alignment. ” IEEE Micro, 37(4), pp. 20 -28, 2017. 14
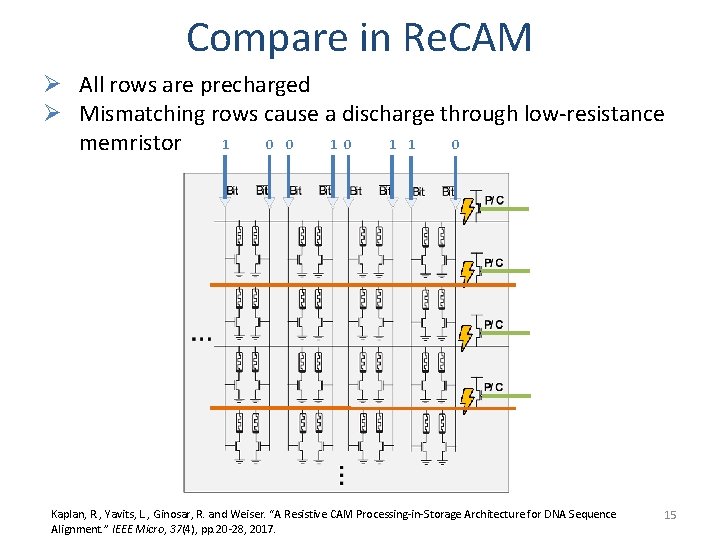
Compare in Re. CAM Ø All rows are precharged Ø Mismatching rows cause a discharge through low-resistance 1 0 0 1 1 memristor Kaplan, R. , Yavits, L. , Ginosar, R. and Weiser. “A Resistive CAM Processing-in-Storage Architecture for DNA Sequence Alignment. ” IEEE Micro, 37(4), pp. 20 -28, 2017. 15
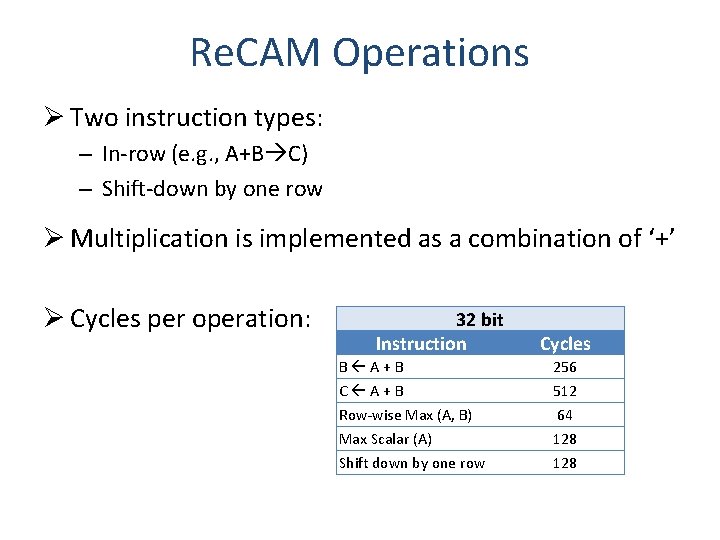
Re. CAM Operations Ø Two instruction types: – In-row (e. g. , A+B C) – Shift-down by one row Ø Multiplication is implemented as a combination of ‘+’ Ø Cycles per operation: 32 bit Instruction B A+B C A+B Row-wise Max (A, B) Max Scalar (A) Shift down by one row Cycles 256 512 64 128
Part 2: Our Contribution, Bio. SEAL 1. 2. 3. 4. Re. CAM drawbacks & our solutions New Re. CAM: Batch Write Re. CAM (BW-Re. CAM) Bio. SEAL Array Performance and energy efficiency comparsions to other architectures
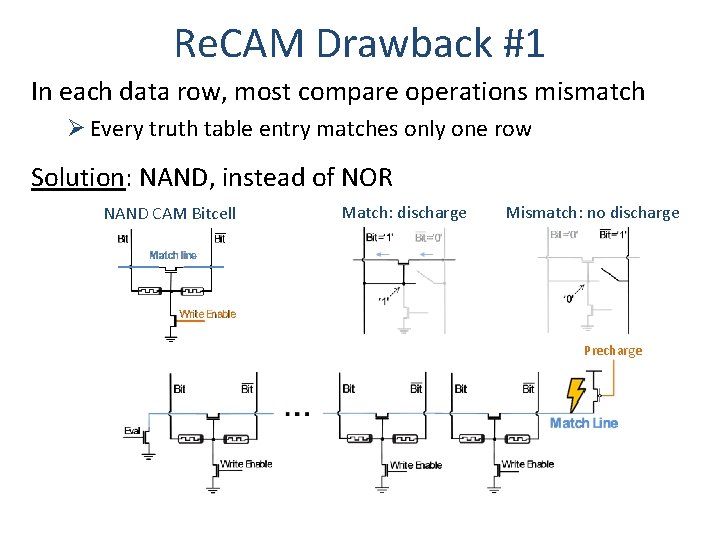
Re. CAM Drawback #1 In each data row, most compare operations mismatch Ø Every truth table entry matches only one row Solution: NAND, instead of NOR NAND CAM Bitcell Match: discharge Mismatch: no discharge Precharge
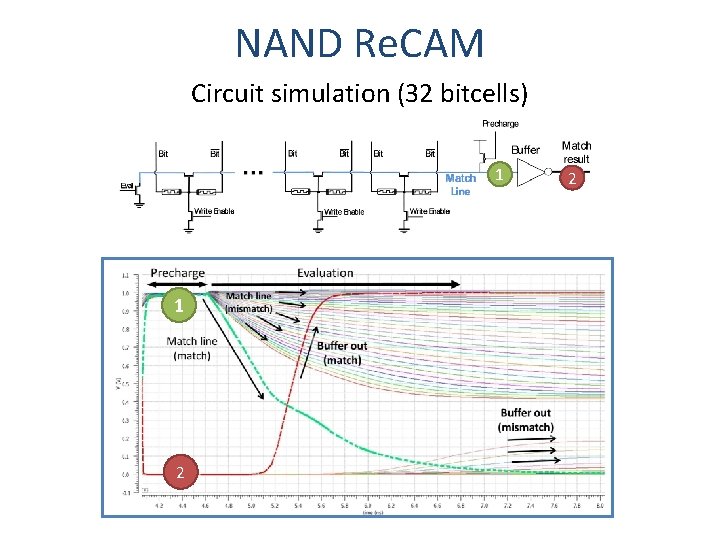
NAND Re. CAM Circuit simulation (32 bitcells) 1 1 2 2
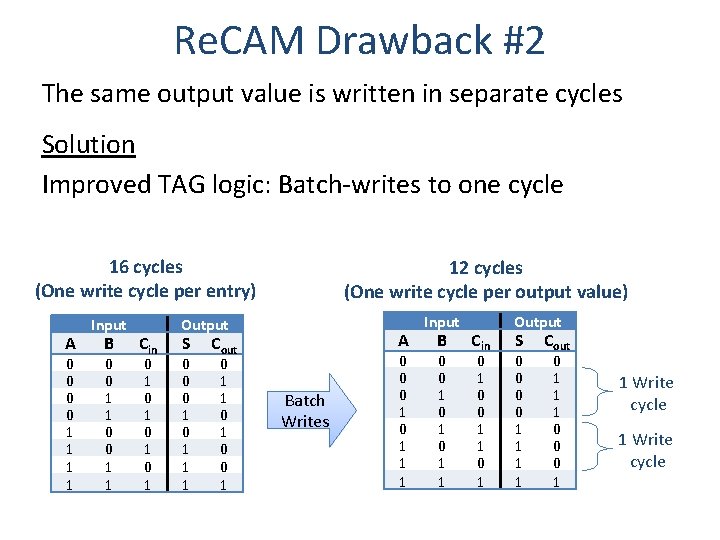
Re. CAM Drawback #2 The same output value is written in separate cycles Solution Improved TAG logic: Batch-writes to one cycle 16 cycles (One write cycle per entry) A 0 0 1 1 Input B 0 0 1 1 Cin 0 1 0 1 12 cycles (One write cycle per output value) Output S 0 0 0 1 1 1 A Cout 0 1 1 0 0 1 Batch Writes 0 0 0 1 1 1 Input B 0 0 1 0 1 1 Cin 0 1 0 0 1 1 0 1 Output S 0 0 1 1 Cout 0 1 1 1 0 0 0 1 1 Write cycle
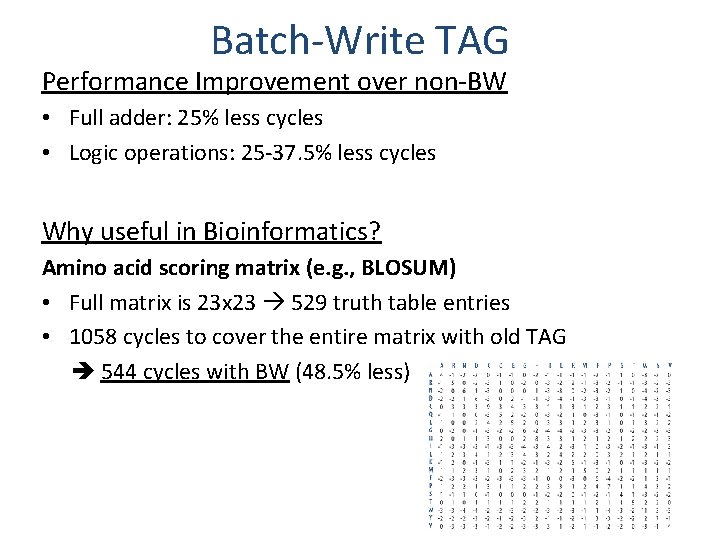
Batch-Write TAG Performance Improvement over non-BW • Full adder: 25% less cycles • Logic operations: 25 -37. 5% less cycles Why useful in Bioinformatics? Amino acid scoring matrix (e. g. , BLOSUM) • Full matrix is 23 x 23 529 truth table entries • 1058 cycles to cover the entire matrix with old TAG 544 cycles with BW (48. 5% less)
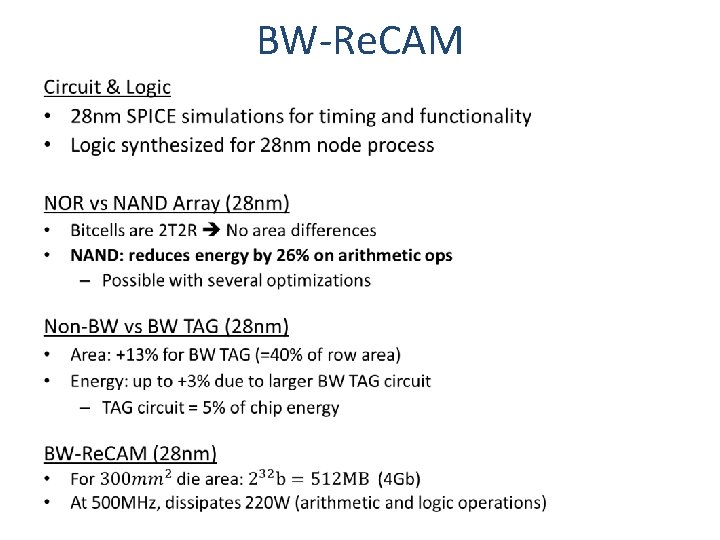
BW-Re. CAM •
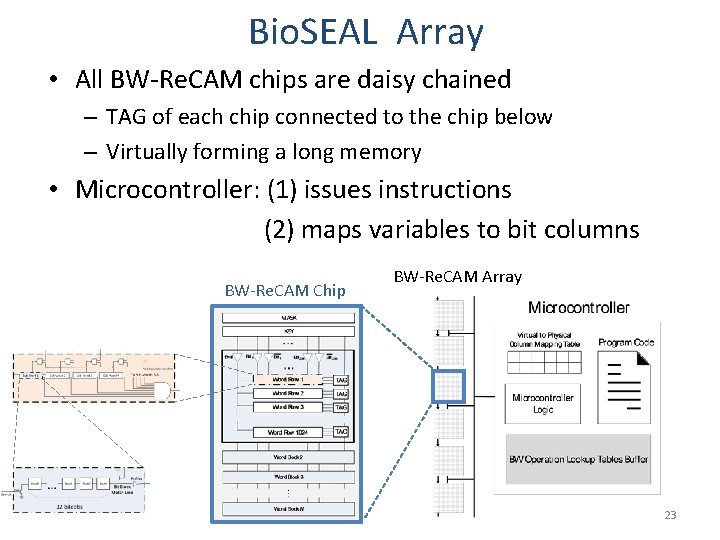
Bio. SEAL Array • All BW-Re. CAM chips are daisy chained – TAG of each chip connected to the chip below – Virtually forming a long memory • Microcontroller: (1) issues instructions (2) maps variables to bit columns BW-Re. CAM Chip BW-Re. CAM Array 23
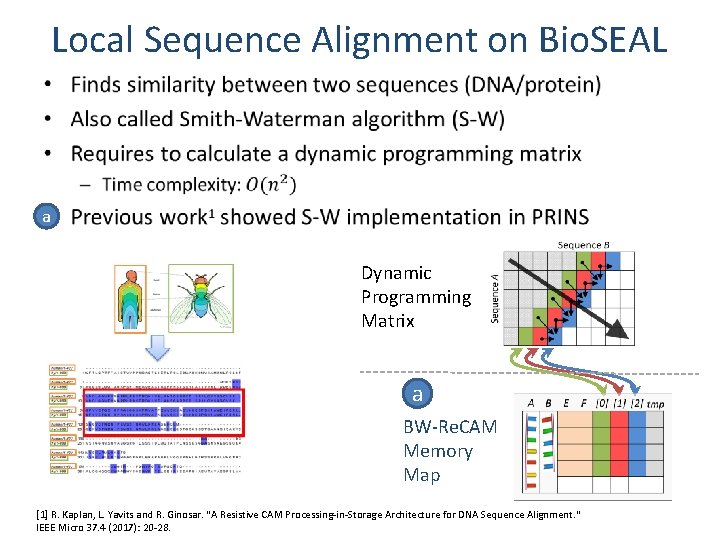
Local Sequence Alignment on Bio. SEAL • a Dynamic Programming Matrix a BW-Re. CAM Memory Map [1] R. Kaplan, L. Yavits and R. Ginosar. "A Resistive CAM Processing-in-Storage Architecture for DNA Sequence Alignment. " IEEE Micro 37. 4 (2017): 20 -28.
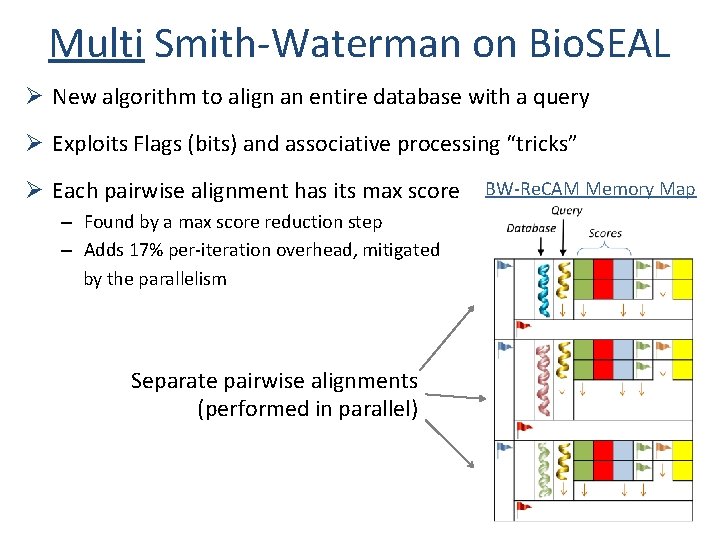
Multi Smith-Waterman on Bio. SEAL Ø New algorithm to align an entire database with a query Ø Exploits Flags (bits) and associative processing “tricks” Ø Each pairwise alignment has its max score – Found by a max score reduction step – Adds 17% per-iteration overhead, mitigated by the parallelism Separate pairwise alignments (performed in parallel) BW-Re. CAM Memory Map
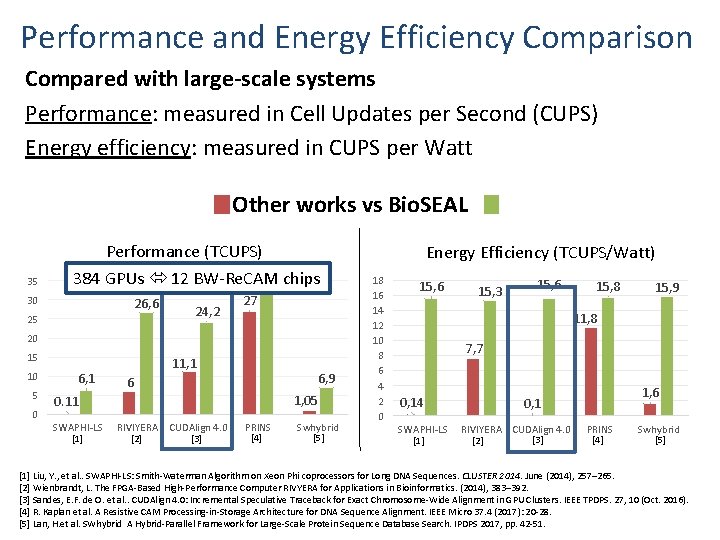
Performance and Energy Efficiency Comparison Compared with large-scale systems Performance: measured in Cell Updates per Second (CUPS) Energy efficiency: measured in CUPS per Watt Other works vs Bio. SEAL 35 Performance (TCUPS) 384 GPUs 12 BW-Re. CAM 31, 8 chips 30 26, 6 25 24, 2 27 20 15 10 5 0 6, 1 11, 1 6, 9 6 1, 05 0. 11 SWAPHI-LS [1] RIVIYERA CUDAlign 4. 0 [3] [2] PRINS [4] Swhybrid [5] Energy Efficiency (TCUPS/Watt) 18 16 14 12 10 8 6 4 2 0 15, 6 15, 3 15, 6 15, 8 15, 9 11, 8 7, 7 0, 14 SWAPHI-LS [1] 1, 6 0, 1 RIVIYERA CUDAlign 4. 0 [3] [2] PRINS [4] Swhybrid [5] [1] Liu, Y. , et al. . SWAPHI-LS: Smith-Waterman Algorithm on Xeon Phi coprocessors for Long DNA Sequences. CLUSTER 2014. June (2014), 257– 265. [2] Wienbrandt, L. The FPGA-Based High-Performance Computer RIVYERA for Applications in Bioinformatics. (2014), 383– 392. [3] Sandes, E. F. de O. et al. . CUDAlign 4. 0: Incremental Speculative Traceback for Exact Chromosome-Wide Alignment in GPU Clusters. IEEE TPDPS. 27, 10 (Oct. 2016). [4] R. Kaplan et al. A Resistive CAM Processing-in-Storage Architecture for DNA Sequence Alignment. IEEE Micro 37. 4 (2017): 20 -28. [5] Lan, H. et al. SWhybrid A Hybrid-Parallel Framework for Large-Scale Protein Sequence Database Search. IPDPS 2017, pp. 42 -51.

Thank you
- Slides: 27