Bio Sci 203 bblecture 5 c DNA library
Bio Sci 203 bb-lecture 5 - c. DNA library screening & sequence characterization • Bruce Blumberg (blumberg@uci. edu) – office – 2113 E Mc. Gaugh Hall – 824 -8573 – lab (x 46873, x 43116) – office hours MWF 11 -12. • • http: //blumberg-serv. bio. uci. edu/bio 203 -w 2002/index. htm http: //blumberg. bio. uci. edu/bio 203 -w 2002/index. htm • This week – c. DNA identification – Protein protein binding assays – Characterization of Selected DNA Sequences • DNA sequence analysis – m. RNA Analysis – Transcript mapping Bio. Sci 203 blumberg lecture 5 page 1 ©copyright Bruce Blumberg 2001 -2005. All rights reserved
How to identify your gene of interest • Screening methods depend on what type of information you have in hand. – Related gene from another species? • Low stringency hybridization – A piece of genomic DNA? • Hybridization – A mutant • Complementation • Positional cloning – A functional assay? • Expression screening – An antibody? • Expression library screening – A partial amino acid sequence? • Oligonucleotide screening – A DNA element required for expression of an interesting gene? • Various binding protein strategies – An interacting protein? • Interaction screening – A specific tissue or embryonic stage? • Subtracted or +/- screening Bio. Sci 203 blumberg lecture 5 page 2 ©copyright Bruce Blumberg 2001 -2005. All rights reserved

How to identify your gene of interest (contd) • Cloning by complementation – generally only useful with manipulable genetic systems • yeast • Drosophila • C. elegans • zebrafish – presumes that complemented mutant is readily observable – Approach • transfer pooled c. DNA libraries in expression vectors into the mutant – or m. RNA pools derived from libraries • assay for rescue • subdivide positive pools and repeat – advantages • direct functional test • rapid compared with chromosome walking – disadvantages • fairly tedious • dependent on library quality • requires easily observable rescue Bio. Sci 203 blumberg lecture 5 page 3 ©copyright Bruce Blumberg 2001 -2005. All rights reserved

How to identify your gene of interest (contd) • Positional cloning – If your mutant results from a transposon insertion then this can be recovered – If insertion is a P-element or gene trap • Make genomic library from mutant – What type of library will you make (λ, BAC, etc)? Why? • Screen with transposon – Recover positives, sequence flanking region • Use flanking sequence to screen normal genomic library – What type of library will you screen (λ, BAC, etc)? Why? – If insertion is a gene trap or related • You can digest mutant DNA with an enzyme that linearizes the vector • Ligate and transform • Colonies that form should have flanking region – sequence • Use this to screen normal library – OR • Use inverse PCR to get flanking sequence from plasmid and use this to probe library Bio. Sci 203 blumberg lecture 5 page 4 ©copyright Bruce Blumberg 2001 -2005. All rights reserved

How to identify your gene of interest (contd) • Functional screening (expression cloning) – if you have a functional assay, expression cloning is reasonable – strategy: • Large pools (~10, 000) of c. DNAs tested for function – microinjection – transfection – receptor binding (panning) • positive pools are subdivided and retested to obtain pure c. DNAs • cycle is repeated until single clones obtained – Advantages • functional approach • in vivo testing is possible • can identify secreted proteins and receptors – Disadvantages • Slow and tedious • sensitivity issue due to pool size • extensive retesting of pools is required – applications: • many receptors and transporters cloned this way Bio. Sci 203 blumberg lecture 5 page 5 ©copyright Bruce Blumberg 2001 -2005. All rights reserved

How to identify your gene of interest (contd) • Antibody screening of c. DNA expression libraries – requirements • antibody must recognize denatured epitope (western blot) – many monoclonals recognize 3 -D or sugar epitopes • affinity purified antibodies work best • c. DNA expression library, e. g. , λgt 11 series – approach • plate library and induce replicate filters • incubate with antibody, wash and develop the filters • repeat until a pure clone is obtained – verification • affinity purify antibody with phage fusion protein – western with original protein – advantages • best choice if only antibody is available – disadvantages • λgt 11 and relatives are painful to work with • your antibody may not be suitable – sugar directed – structural epitope Bio. Sci 203 blumberg lecture 5 page 6 ©copyright Bruce Blumberg 2001 -2005. All rights reserved

How to identify your gene of interest (contd) • A partial amino acid sequence? – Purified protein and have one or more partial amino acid sequences • make a peptide antibody and screen (slow) • Oligonucleotide screening based on aa sequence – multiple codons for most aa • PCR between multiple primers – three types of oligos in use • long guess-mers - pick the wobble base – relies on low stringency hybridization • inosine - use inosine for multiple bases – I: C >> others • degenerate oligos (mixtures of all possible seqs) – mixtures of < 1024 virtually always work Bio. Sci 203 blumberg lecture 5 page 7 ©copyright Bruce Blumberg 2001 -2005. All rights reserved
How to identify your gene of interest (contd) • A partial amino acid sequence (contd)? – approach • pick an aa sequence that predicts a reasonable probe complexity (~1024 fold)(avoid ser, leu, arg) WHY? • synthesize fully degenerate mixture of probes and label • hybridize at low stringency (Tm-25 for the most AT rich sequence) • wash at high stringency in 3 M tetramethylammonium chloride – TMAC stabilizes AT base pairs A-T = G-C – Tm is a strict function of length – works best for 21 -23 mers – degenerate oligo and TMAC • advantages – degenerate oligos always work – fast – only requires a single sequence • disadvantages – TMAC method requires strict adherence to technique – aa sequence may not predict a good oligo » e. g. , too many leu, ser or arg Bio. Sci 203 blumberg lecture 5 page 8 ©copyright Bruce Blumberg 2001 -2005. All rights reserved

How to identify your gene of interest (contd) • A partial amino acid sequence (contd) – PCR – design primers to two conserved sequences, amplify, clone • advantages – very fast – almost anyone can manage • disadvantages – requires 2 good sequences – PCR errors may give incorrect sequence Bio. Sci 203 blumberg lecture 5 page 9 ©copyright Bruce Blumberg 2001 -2005. All rights reserved
How to identify your gene of interest (contd) • A DNA element required for expression of an interesting gene? – How to identify what factors bind to putative elements? • examine the sequence – does it contain known binding sites? – Check TRANSFAC database » http: //www. gene-regulation. com/ – if yes, do such proteins bind to the isolated element in gel-shift experiments? • do the elements bind proteins from nuclear extracts? – gel shift (EMSA) experiments • clone the elements into reporters with minimal promoters. – do these constructs recapitulate activity? – What does the sequence tell you about the binding protein? • AGGTCATGACCT Dyad symmetry always means multimeric protein No symmetry usually means monomeric protein Bio. Sci 203 blumberg lecture 5 page 10 ©copyright Bruce Blumberg 2001 -2005. All rights reserved
How to identify your gene of interest (contd) • Biochemical purification of binding proteins – tedious, considerable biochemical skill required – two basic approaches • fractionate nuclear extracts chromatographically and test fractions for ability to bind the element • DNA-affinity chromatography – multimerize the element and bind to a resin – pass nuclear extracts across column and purify specific binding proteins – protein microsequencing – predict DNA sequence from amino acid sequence • look in the database • prepare oligonucleotides and screen library Bio. Sci 203 blumberg lecture 5 page 11 ©copyright Bruce Blumberg 2001 -2005. All rights reserved
How to identify your gene of interest (contd) • Biochemical purification of binding proteins (contd) – advantages • gold standard • if you can purify proteins, this will always work – not so many good protein biochemists • works for dimeric proteins and complexes – disadvantages • slow, tedious • need good protein sequencing facility • biochemical expertise required • expense of preparing preparative quantities of nuclear extracts Bio. Sci 203 blumberg lecture 5 page 12 ©copyright Bruce Blumberg 2001 -2005. All rights reserved

How to identify your gene of interest (contd) • Molecular biological approaches to identifying binding proteins – oligonucleotide screening of expression libraries (Singh screening) • multimerize oligonucleotide and label with 32 P • screen expression library to identify binding proteins • advantages – straightforward – much less biochemical expertise required than biochemical purification – relatively fast • disadvantages – can’t detect binding if multiple partners are required – fair amount of “touch” required Bio. Sci 203 blumberg lecture 5 page 13 ©copyright Bruce Blumberg 2001 -2005. All rights reserved
How to identify your gene of interest (contd)) • Molecular biological approaches to identifying binding proteins – expression cloning (sib screening) • clone element of interest (or promoter) into a suitable reporter construct (e. g. luciferase) • transfect (or inject, or infect, etc) pools (~10, 000 c. DNAs each) of c. DNA expression libraries and assay for reporter gene • retest positive pools in smaller aliquots (~1000) • repeat until a pure c. DNA is found – advantages • functional approach • presumably using the appropriate cell type so modifications occur • possibility to detect dimers with endogenous proteins – disadvantages • VERY TEDIOUS • very slow, much duplication in pools, extensive rescreening is required • could be expensive Bio. Sci 203 blumberg lecture 5 page 14 ©copyright Bruce Blumberg 2001 -2005. All rights reserved
How to identify your gene of interest (contd) • Molecular biological approaches to identifying binding proteins – in vitro expression cloning (IVEC) • transcribe and translate c. DNA libraries in vitro into small pools of proteins (~100) • EMSA to test protein pools for element binding • unpool c. DNAs and retest • advantages – functional approach – smaller pools increase sensitivity • disadvantages – can’t detect dimers – very expensive (TNT lysate) – considerable rescreening still required – tedious, countless DNA minipreps required Bio. Sci 203 blumberg lecture 5 page 15 ©copyright Bruce Blumberg 2001 -2005. All rights reserved
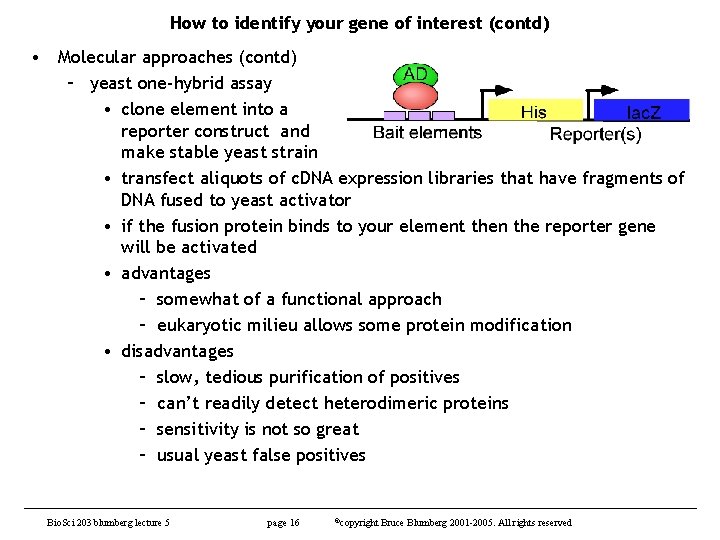
How to identify your gene of interest (contd) • Molecular approaches (contd) – yeast one-hybrid assay • clone element into a reporter construct and make stable yeast strain • transfect aliquots of c. DNA expression libraries that have fragments of DNA fused to yeast activator • if the fusion protein binds to your element then the reporter gene will be activated • advantages – somewhat of a functional approach – eukaryotic milieu allows some protein modification • disadvantages – slow, tedious purification of positives – can’t readily detect heterodimeric proteins – sensitivity is not so great – usual yeast false positives Bio. Sci 203 blumberg lecture 5 page 16 ©copyright Bruce Blumberg 2001 -2005. All rights reserved

How to identify your gene of interest (contd) • You have one protein and want to identify proteins that interact with it – some sort of interaction screen is indicated • straight biochemistry • phage display • two hybrid • in vitro expression cloning Bio. Sci 203 blumberg lecture 5 page 17 ©copyright Bruce Blumberg 2001 -2005. All rights reserved
How to identify your gene of interest (contd) • biochemical approach – purify cellular proteins that interact with your protein • co-immunoprecipitation • affinity chromatography • biochemical fractionation – pure protein(s) are microsequenced • if not in database then make oligonucleotides and screen c. DNA library from appropriate tissues – advantage • functional approach • stringency can be manipulated • can identify multimeric proteins or complexes • will work if you can purify proteins – disadvantages • much skill required • low throughput • considerable optimization required Bio. Sci 203 blumberg lecture 5 page 18 ©copyright Bruce Blumberg 2001 -2005. All rights reserved
- Slides: 18