Bio PAX Biological Pathways Data Exchange www biopaxwiki

Bio. PAX Biological Pathways Data Exchange www. biopaxwiki. org Joanne Luciano, Ph. D University of Manchester, Harvard Medical School Bio. Pathways Consortium, Bio. PAX Group, Predictive Medicine, Inc. 16/10/2021 25 Jan 2006 Cambridge, MA USA 1

Pathway Data Why does HCLS care? (where we fit) Pathway Research has Broad Impact – – Drug Discovery (pathway of target, safety) Basic Science (identify pathways) Disease Research (cancer pathways, diabetes, malaria) Environmental Research (microbial research) Combine knowledge from multiple sources – Whole is greater than the sum of its parts – Biological knowledge is fragmented and isolated – Need database to manage resources 16/10/2021 2

What is a Pathway? Depends on who you ask! Glycolysis Protein-Protein Apoptosis TFs in E. coli Metabolic Pathways Molecular Interaction Networks Signaling Pathways Gene Regulatory Networks 16/10/2021 3
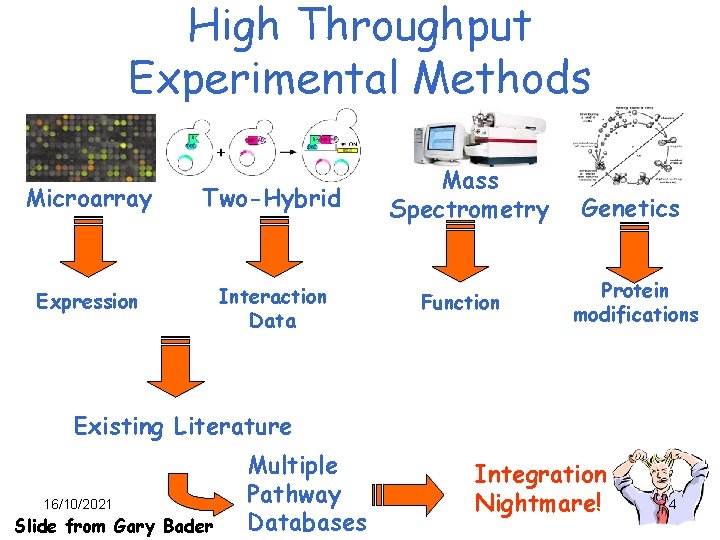
High Throughput Experimental Methods Microarray Two-Hybrid Expression Interaction Data Mass Spectrometry Function Genetics Protein modifications Existing Literature 16/10/2021 Slide from Gary Bader Multiple Pathway Databases Integration Nightmare! 4
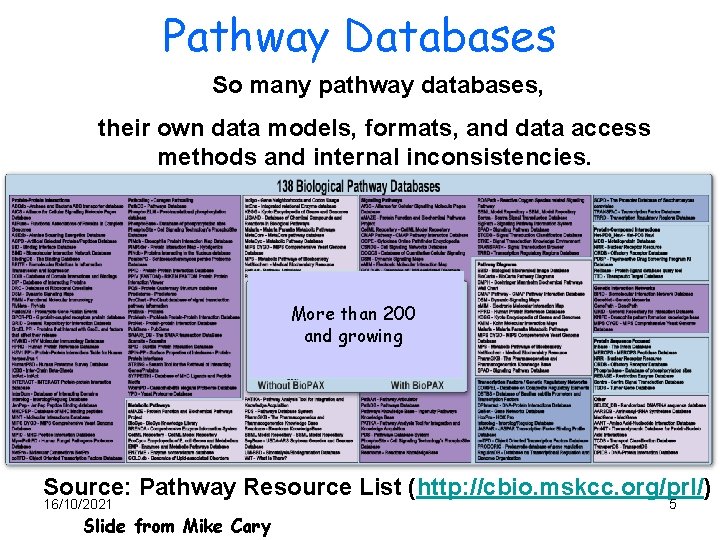
Pathway Databases So many pathway databases, their own data models, formats, and data access methods and internal inconsistencies. More than 200 and growing Source: Pathway Resource List (http: //cbio. mskcc. org/prl/) 16/10/2021 Slide from Mike Cary 5
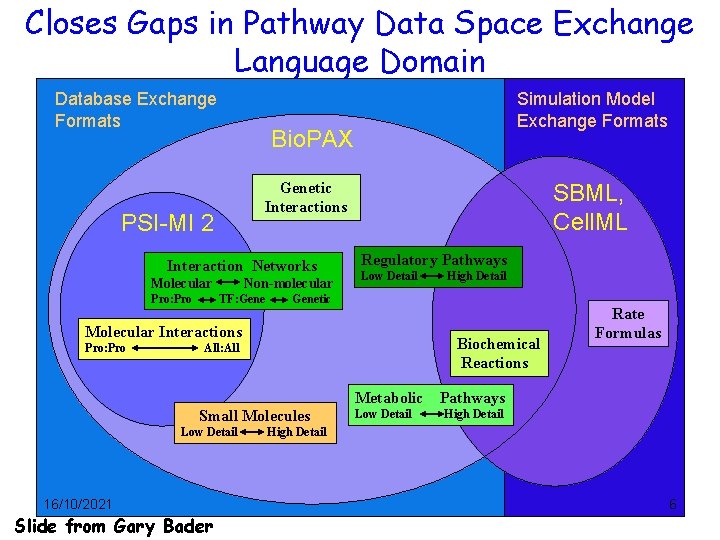
Closes Gaps in Pathway Data Space Exchange Language Domain Database Exchange Formats Bio. PAX Genetic Interactions PSI-MI 2 Interaction Networks Molecular Pro: Pro Simulation Model Exchange Formats Non-molecular TF: Gene SBML, Cell. ML Regulatory Pathways Low Detail Genetic Molecular Interactions Pro: Pro High Detail Biochemical Reactions All: All Rate Formulas Metabolic Pathways Small Molecules Low Detail 16/10/2021 Slide from Gary Bader Low Detail High Detail 6
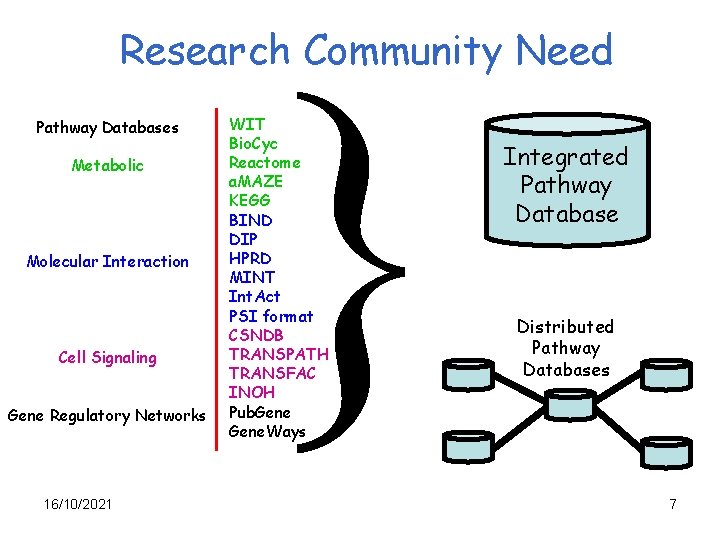
} Research Community Need Pathway Databases Metabolic Molecular Interaction Cell Signaling Gene Regulatory Networks 16/10/2021 WIT Bio. Cyc Reactome a. MAZE KEGG BIND DIP HPRD MINT Int. Act PSI format CSNDB TRANSPATH TRANSFAC INOH Pub. Gene. Ways Integrated Pathway Database Distributed Pathway Databases 7
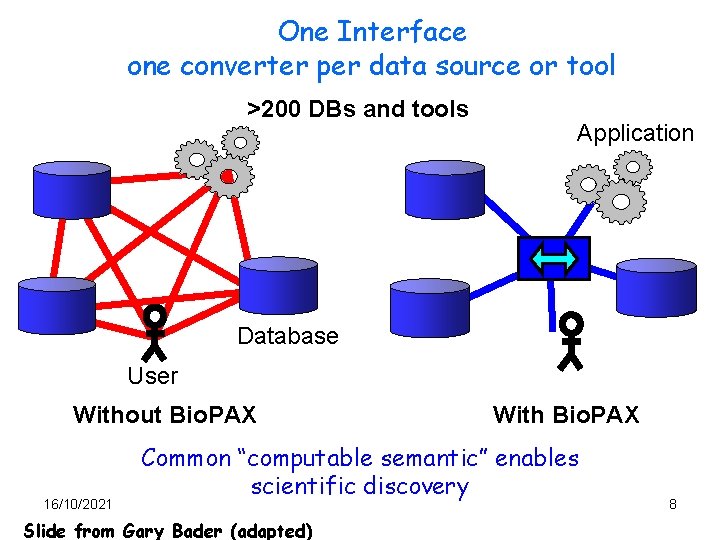
One Interface one converter per data source or tool >200 DBs and tools Application Database User Without Bio. PAX 16/10/2021 With Bio. PAX Common “computable semantic” enables scientific discovery Slide from Gary Bader (adapted) 8

Design Goals Encapsulation – An entire pathway in one record Compatible – Use existing standards wherever possible Computable – From file reading to logical inference Successful – Buy-in from the research community 16/10/2021 9

Why OWL DL? Expressivity (biology = “complex relationships” • W 3 C Standard (use existing (and upcoming) standards) “Semantic Web enabled” • OWL has representations in RDF and XML (XML the exchange language) Machine Computable Enable full reasoning capability from file reading to logical inference – facilitate integration of knowledge, data, tool development – uncover inconsistencies and new knowledge 16/10/2021 10
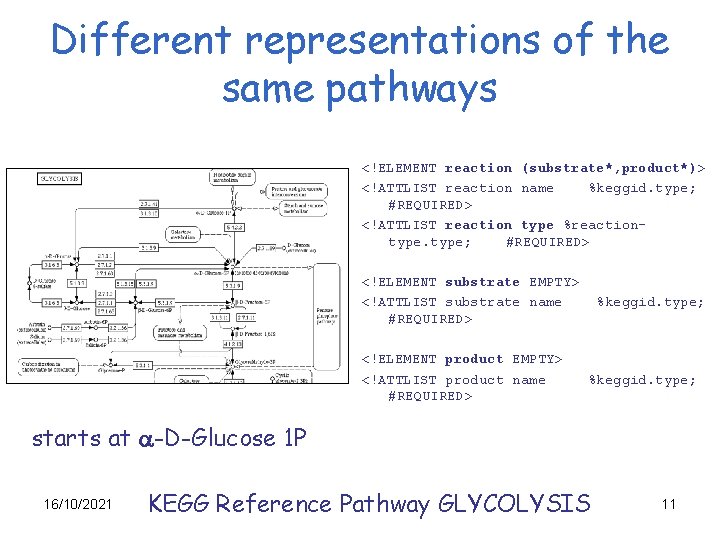
Different representations of the same pathways <!ELEMENT reaction (substrate*, product*)> <!ATTLIST reaction name %keggid. type; #REQUIRED> <!ATTLIST reaction type %reactiontype; #REQUIRED> <!ELEMENT substrate EMPTY> <!ATTLIST substrate name #REQUIRED> <!ELEMENT product EMPTY> <!ATTLIST product name #REQUIRED> %keggid. type; starts at a-D-Glucose 1 P 16/10/2021 KEGG Reference Pathway GLYCOLYSIS 11
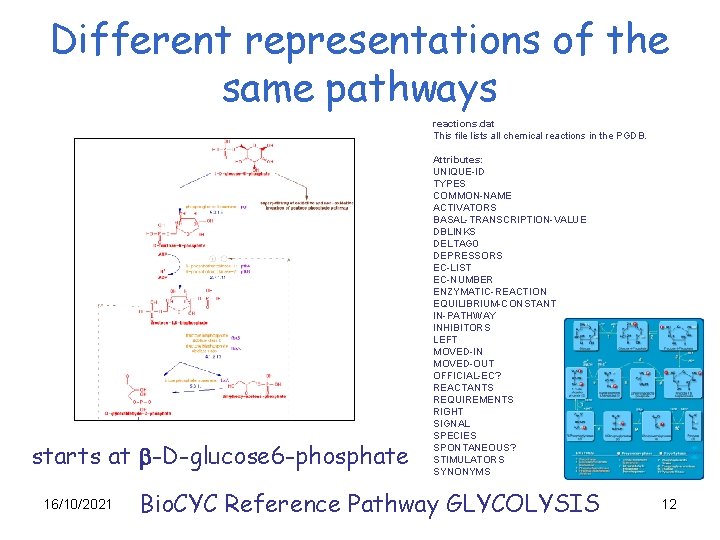
Different representations of the same pathways reactions. dat This file lists all chemical reactions in the PGDB. starts at b-D-glucose 6 -phosphate 16/10/2021 Attributes: UNIQUE-ID TYPES COMMON-NAME ACTIVATORS BASAL-TRANSCRIPTION-VALUE DBLINKS DELTAG 0 DEPRESSORS EC-LIST EC-NUMBER ENZYMATIC-REACTION EQUILIBRIUM-CONSTANT IN-PATHWAY INHIBITORS LEFT MOVED-IN MOVED-OUT OFFICIAL-EC? REACTANTS REQUIREMENTS RIGHT SIGNAL SPECIES SPONTANEOUS? STIMULATORS SYNONYMS Bio. CYC Reference Pathway GLYCOLYSIS 12

Bio. PAX uses other ontologies • Use pointers to existing ontologies to provide supplemental annotation where appropriate – Cellular location GO Component – Cell type Cell. obo – Organism NCBI taxon DB • Incorporate other standards where appropriate – Chemical structure SMILES, CML, In. Ch. I 16/10/2021 13
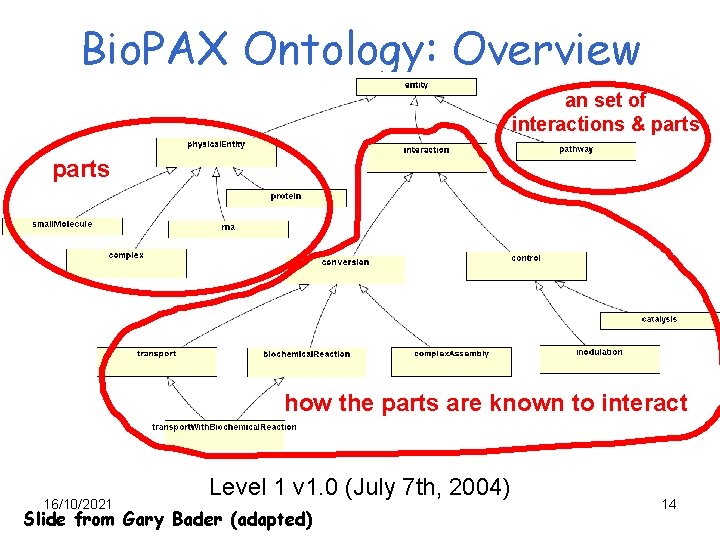
Bio. PAX Ontology: Overview an set of interactions & parts how the parts are known to interact 16/10/2021 Level 1 v 1. 0 (July 7 th, 2004) Slide from Gary Bader (adapted) 14
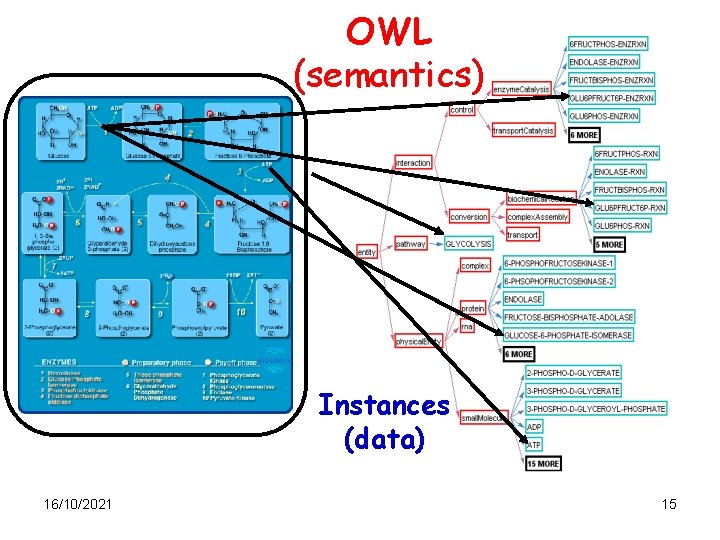
OWL (semantics) Instances (data) 16/10/2021 15
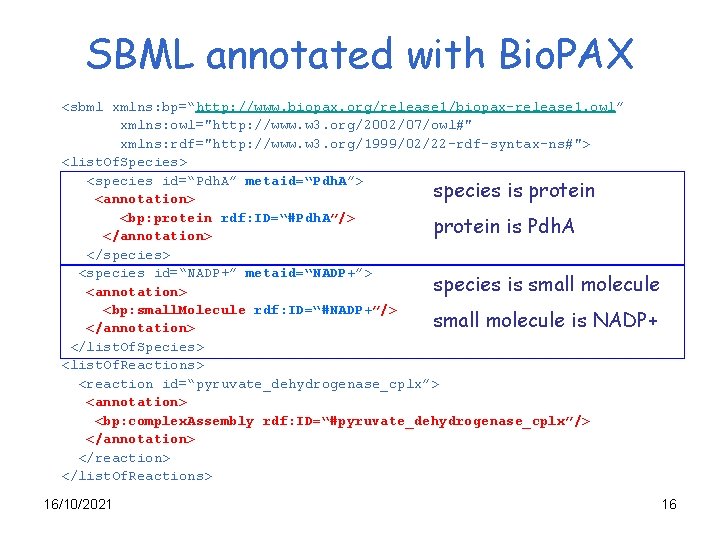
SBML annotated with Bio. PAX <sbml xmlns: bp=“http: //www. biopax. org/release 1/biopax-release 1. owl” xmlns: owl="http: //www. w 3. org/2002/07/owl#" xmlns: rdf="http: //www. w 3. org/1999/02/22 -rdf-syntax-ns#"> <list. Of. Species> <species id=“Pdh. A” metaid=“Pdh. A”> species is protein <annotation> <bp: protein rdf: ID=“#Pdh. A”/> protein is Pdh. A </annotation> </species> <species id=“NADP+” metaid=“NADP+”> species is small molecule <annotation> <bp: small. Molecule rdf: ID=“#NADP+”/> small molecule is NADP+ </annotation> </list. Of. Species> <list. Of. Reactions> <reaction id=“pyruvate_dehydrogenase_cplx”> <annotation> <bp: complex. Assembly rdf: ID=“#pyruvate_dehydrogenase_cplx”/> </annotation> </reaction> </list. Of. Reactions> 16/10/2021 16

Bio. PAX: External References <species id=“pyruvate” metaid=“pyruvate”> <annotation xmlns: bp=“http: //biopax. org/release 1/biopaxrelease 1. owl”> <bp: small. Molecule rdf: ID=“#pyruvate”> <bp: Xref> <bp: unification. Xref rdf: ID=“#unification. Xref 119"> <bp: DB>LIGAND</bp: DB> <bp: ID>c 00022</bp: ID> </bp: unification. Xref> </bp: small. Molecule> </annotation> 16/10/2021 17 </species>

Bio. PAX: Synonyms <species id=“pyruvate” metaid=“pyruvate”> <annotation xmlns: bp=“http: //biopax. org/release 1/biopax_release 1. owl”/> <bp: small. Molecule rdf: ID=“#pyruvate” > <bp: SYNONYMS>2 -oxo-propionic acid</bp: SYNONYMS> <bp: SYNONYMS>2 -oxopropanoate</bp: SYNONYMS> <bp: SYNONYMS>BTS</bp: SYNONYMS> <bp: SYNONYMS>pyruvic acid</bp: SYNONYMS> </bp: small. Molecule> </annotation> </species> 16/10/2021 18
Tools Protégé Ontology Editor GKB Editor SRI SWOOP Pellet Racer Fact++ Pathway Tools Edit. Plus (Text editor) Want More: See Jeremy & Alan 16/10/2021 19

Overlap? Integration – Combine sources in a meaningful way Identity – Recognize same things in different contexts and different names Composition – Re-usable representations of composite pathway components • to help us manage, query, and reference Exchange – Agreement on: • What is to be exchanged • How to represent it • How to interpret it Want more? See Alan, Jeremy, me 16/10/2021 20
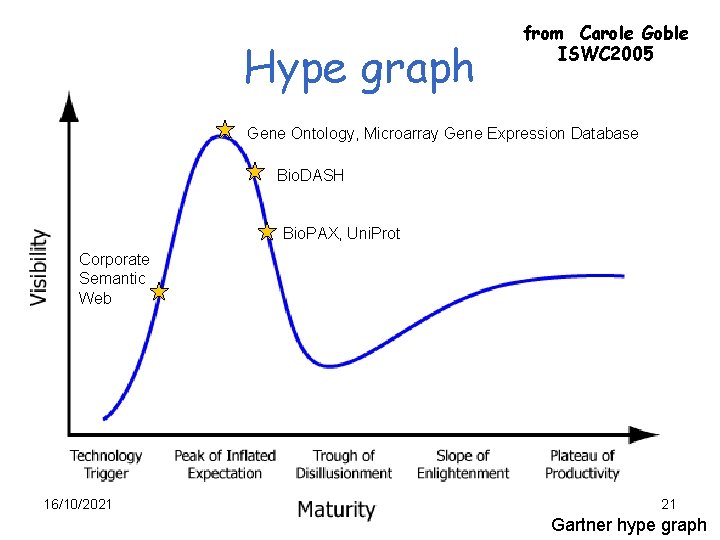
Hype graph from Carole Goble ISWC 2005 Gene Ontology, Microarray Gene Expression Database Bio. DASH Bio. PAX, Uni. Prot Corporate Semantic Web 16/10/2021 21 Gartner hype graph
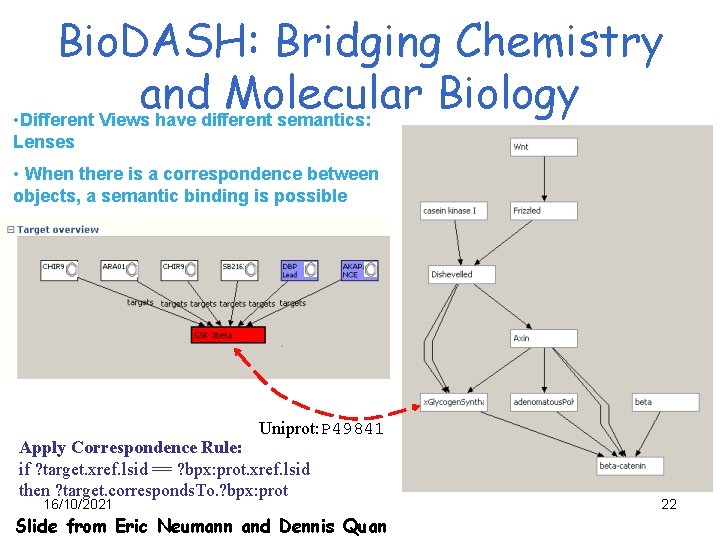
Bio. DASH: Bridging Chemistry and Molecular Biology • Different Views have different semantics: Lenses • When there is a correspondence between objects, a semantic binding is possible Uniprot: P 49841 Apply Correspondence Rule: if ? target. xref. lsid == ? bpx: prot. xref. lsid then ? target. corresponds. To. ? bpx: prot 16/10/2021 Slide from Eric Neumann and Dennis Quan 22
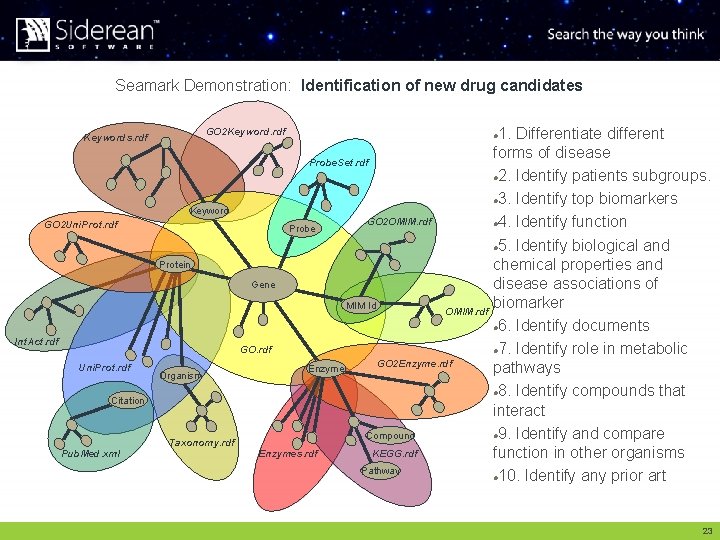
Seamark Demonstration: Identification of new drug candidates Keyword GO 2 Uni. Prot. rdf Protein Int. Act. rdf Uni. Prot. rdf Organism Citation Taxonomy. rdf Pub. Med. xml 1. Differentiate different forms of disease Probe. Set. rdf l 2. Identify patients subgroups. l 3. Identify top biomarkers GO 2 OMIM. rdf l 4. Identify function Probe l 5. Identify biological and chemical properties and Gene disease associations of biomarker MIM Id OMIM. rdf l 6. Identify documents GO. rdf l 7. Identify role in metabolic GO 2 Enzyme. rdf Enzyme pathways l 8. Identify compounds that interact l 9. Identify and compare Compound Enzymes. rdf KEGG. rdf function in other organisms Pathway l 10. Identify any prior art GO 2 Keyword. rdf Keywords. rdf l 23
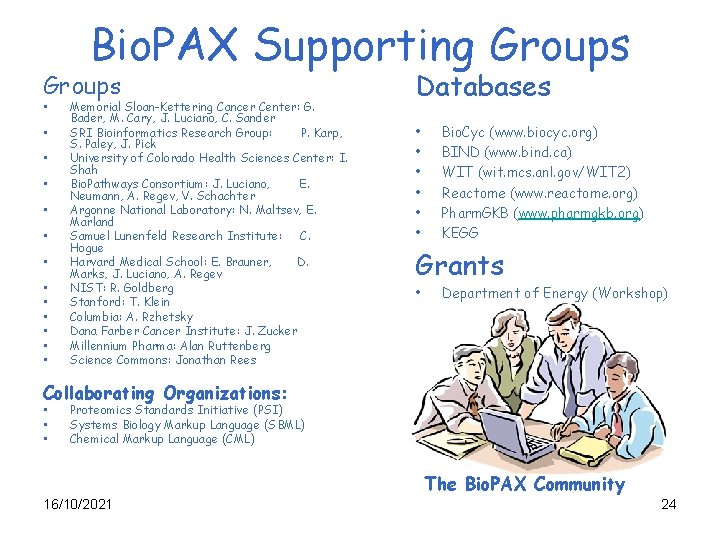
Bio. PAX Supporting Groups Databases • • • • • Memorial Sloan-Kettering Cancer Center: G. Bader, M. Cary, J. Luciano, C. Sander SRI Bioinformatics Research Group: P. Karp, S. Paley, J. Pick University of Colorado Health Sciences Center: I. Shah Bio. Pathways Consortium: J. Luciano, E. Neumann, A. Regev, V. Schachter Argonne National Laboratory: N. Maltsev, E. Marland Samuel Lunenfeld Research Institute: C. Hogue Harvard Medical School: E. Brauner, D. Marks, J. Luciano, A. Regev NIST: R. Goldberg Stanford: T. Klein Columbia: A. Rzhetsky Dana Farber Cancer Institute: J. Zucker Millennium Pharma: Alan Ruttenberg Science Commons: Jonathan Rees Bio. Cyc (www. biocyc. org) BIND (www. bind. ca) WIT (wit. mcs. anl. gov/WIT 2) Reactome (www. reactome. org) Pharm. GKB (www. pharmgkb. org) KEGG Grants • Department of Energy (Workshop) Collaborating Organizations: • • • Proteomics Standards Initiative (PSI) Systems Biology Markup Language (SBML) Chemical Markup Language (CML) The Bio. PAX Community 16/10/2021 24
10/16/2021 25
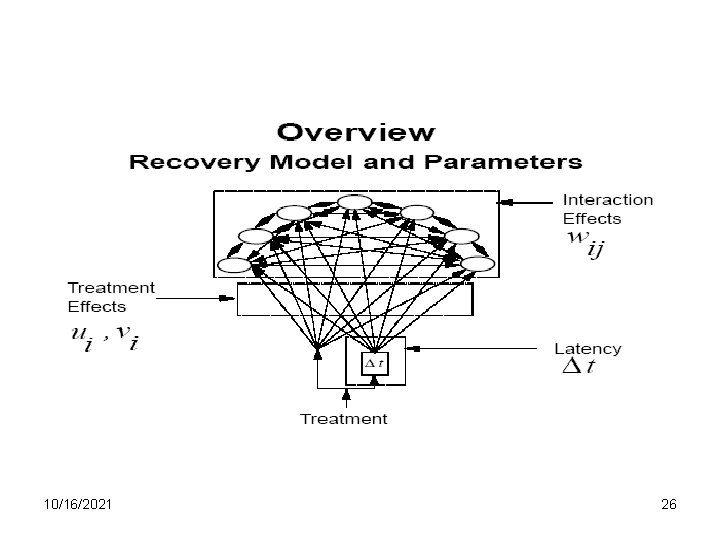
10/16/2021 26
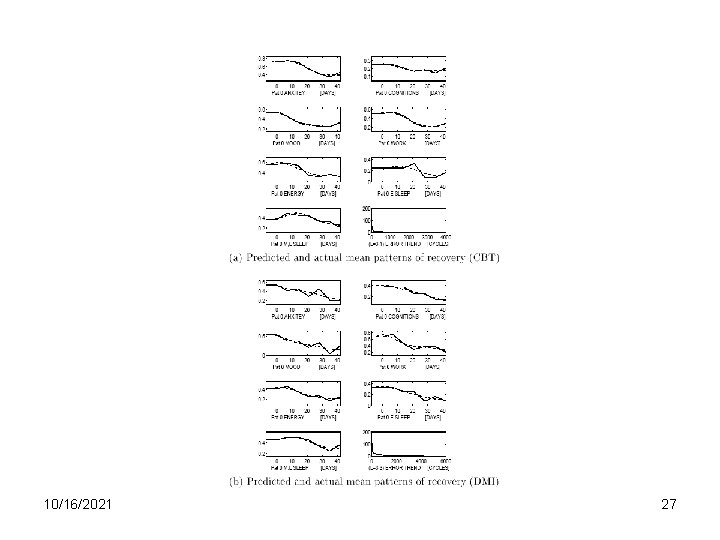
10/16/2021 27
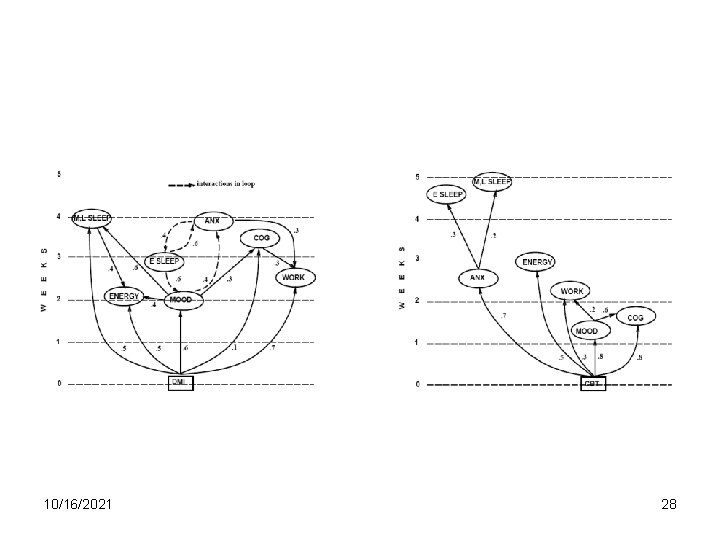
10/16/2021 28
- Slides: 28