BIO 3102 Molecular Evolution Lecture 2 Mutation Prof

BIO 3102 Molecular Evolution Lecture 2. Mutation Prof. Xuhua Xia Department of Biology University of Ottawa xxia@uottawa. ca http: //dambe. bio. uottawa. ca
Outline • Types of mutation classified in different ways: – point mutation, deletion, insertion, inversion, translocation – advantageous, deleterious, neutral – transition, transversion, synonymous, nonsynonymous • Mutation rate and genome size: Constant genomic mutation rate hypothesis • Mutation bias and codon usage • Mutation and substitution • Substitutions as ticks in a clock • Mutation reveals the connection between genotype and phenotype; mutation and genetic diseases • DNA methylation and mutation

Evolution of a DNA sequence: a history of errors and damage Replication and repair of DNA by DNA polymerases are not perfect: • Point mutations (misincorporation of wrong nucleotides • Deletion and insertions of a piece of sequence • Inversion • Translocation • . . .

Mutations - any detectable change in DNA sequence eg. errors in DNA replication/repair - inherited ones of interest in evolutionary studies Deleterious - reduce fitness and will be selected against by purifying selection) Advantageous - increase fitness and have an increased chance of being fixed in the population by natural selection Neutral - will have little effect on phenotype - may be fixed in population by genetic drift
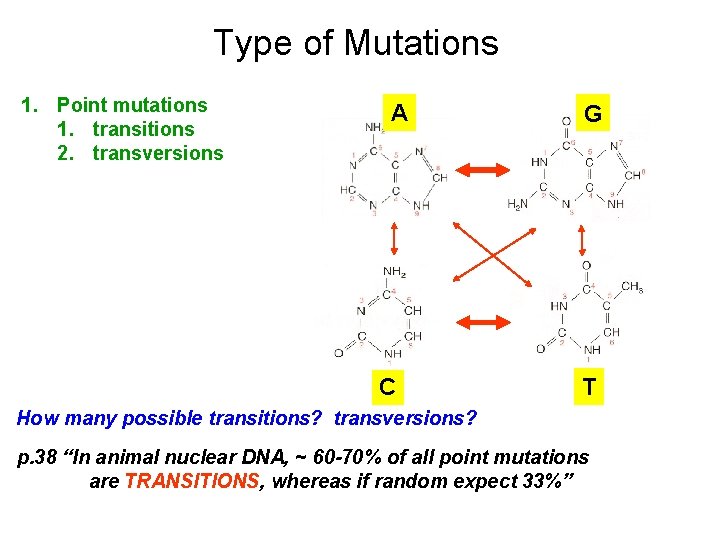
Type of Mutations 1. Point mutations 1. transitions 2. transversions A C G T How many possible transitions? transversions? p. 38 “In animal nuclear DNA, ~ 60 -70% of all point mutations are TRANSITIONS, whereas if random expect 33%”
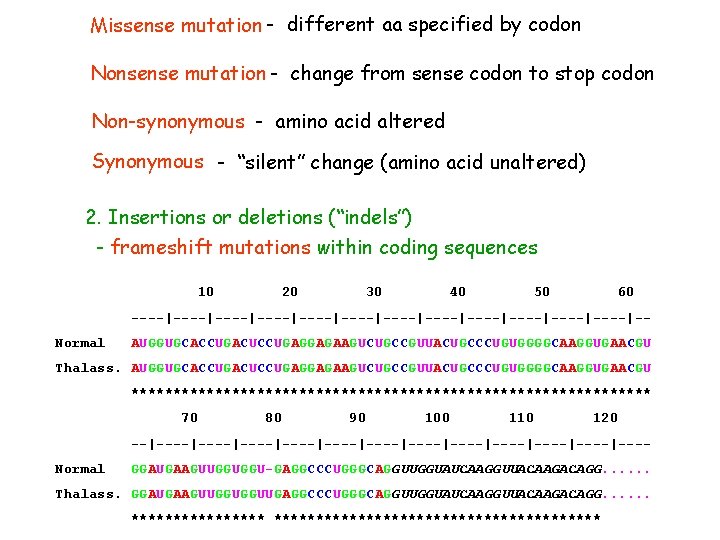
Missense mutation - different aa specified by codon Nonsense mutation - change from sense codon to stop codon Non-synonymous - amino acid altered Synonymous - “silent” change (amino acid unaltered) 2. Insertions or deletions (“indels”) - frameshift mutations within coding sequences 10 20 30 40 50 60 ----|----|----|----|----|----|-Normal AUGGUGCACCUGACUCCUGAGGAGAAGUCUGCCGUUACUGCCCUGUGGGGCAAGGUGAACGU Thalass. AUGGUGCACCUGACUCCUGAGGAGAAGUCUGCCGUUACUGCCCUGUGGGGCAAGGUGAACGU ******************************* 70 80 90 100 110 120 --|----|----|----|----|----|---Normal GGAUGAAGUUGGUGGU-GAGGCCCUGGGCAGGUUGGUAUCAAGGUUACAAGACAGG. . . Thalass. GGAUGAAGUUGGUGGUUGAGGCCCUGGGCAGGUUGGUAUCAAGGUUACAAGACAGG. . . ********************
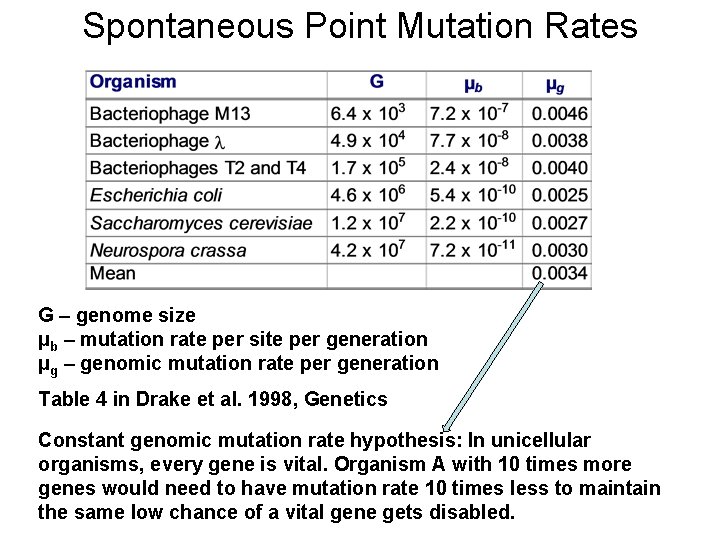
Spontaneous Point Mutation Rates G – genome size µb – mutation rate per site per generation µg – genomic mutation rate per generation Table 4 in Drake et al. 1998, Genetics Constant genomic mutation rate hypothesis: In unicellular organisms, every gene is vital. Organism A with 10 times more genes would need to have mutation rate 10 times less to maintain the same low chance of a vital gene gets disabled.
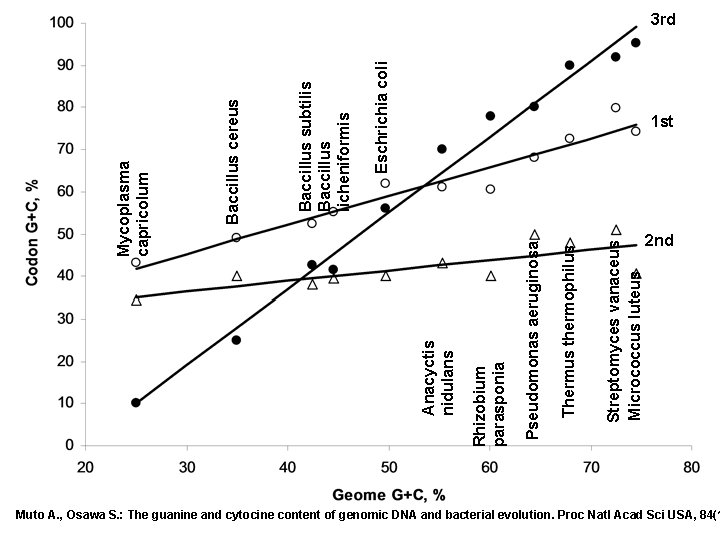
Streptomyces vanaceus Micrococcus luteus Thermus thermophilus Pseudomonas aeruginosa Rhizobium parasponia Anacyctis nidulans Eschrichia coli Baccillus subtilis Baccillus licheniformis Baccillus cereus Mycoplasma capricolum 3 rd 1 st 2 nd Muto A. , Osawa S. : The guanine and cytocine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci USA, 84(1

Mutation, substitution and dating AAA CCC CGG GGC CCC TAT TTT TTG AAG CCC CGG GGC CCC TAT TTG AAA CCC CGG GGC CCC TAT TTT AAG CCT CGG GGC CCC TAT TTG AAT CTC CGG GGC CCC TAT TTT AAG CCT CGG GGC CCT TAT TTG AAT CTC CGG GGC CTC TAT TTT
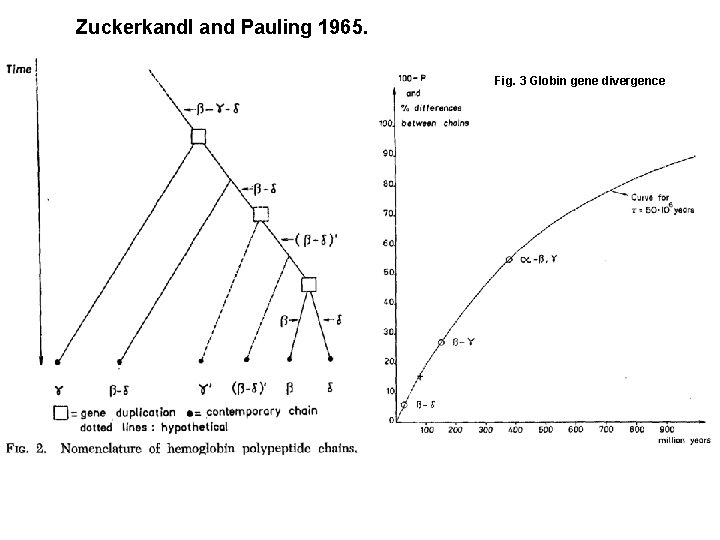
Zuckerkandl and Pauling 1965. Fig. 3 Globin gene divergence
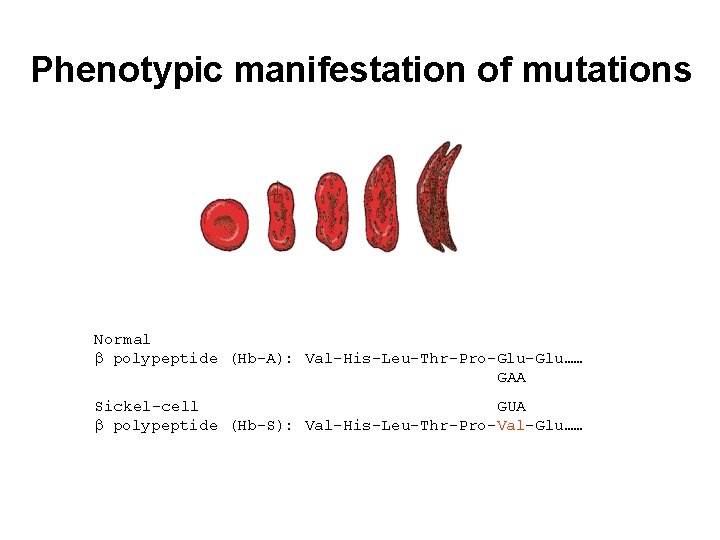
Phenotypic manifestation of mutations Normal polypeptide (Hb-A): Val-His-Leu-Thr-Pro-Glu…… GAA Sickel-cell GUA polypeptide (Hb-S): Val-His-Leu-Thr-Pro-Val-Glu……
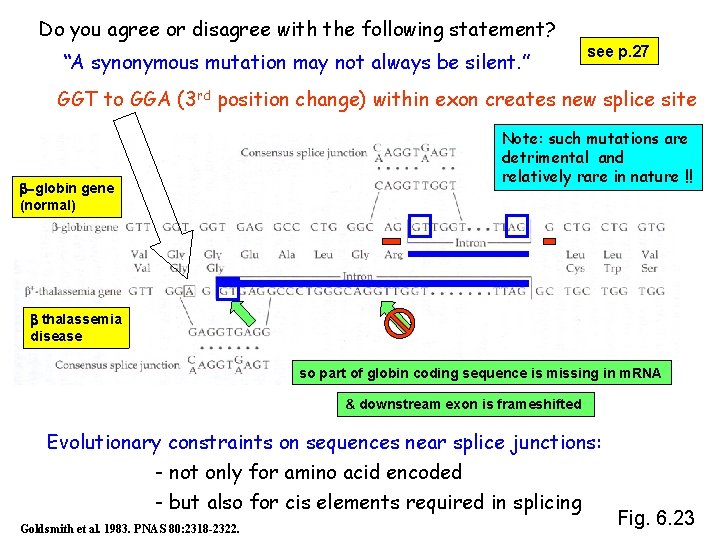
Do you agree or disagree with the following statement? “A synonymous mutation may not always be silent. ” see p. 27 GGT to GGA (3 rd position change) within exon creates new splice site Note: such mutations are detrimental and relatively rare in nature !! b-globin gene (normal) b thalassemia disease so part of globin coding sequence is missing in m. RNA & downstream exon is frameshifted Evolutionary constraints on sequences near splice junctions: - not only for amino acid encoded - but also for cis elements required in splicing Goldsmith et al. 1983. PNAS 80: 2318 -2322. Fig. 6. 23

LP is a derived trait Single origin or multiple independent origins? Lactase persistence (LP) Segurel, L and Bon C. 2017. Ann Rev 18: 297 -319
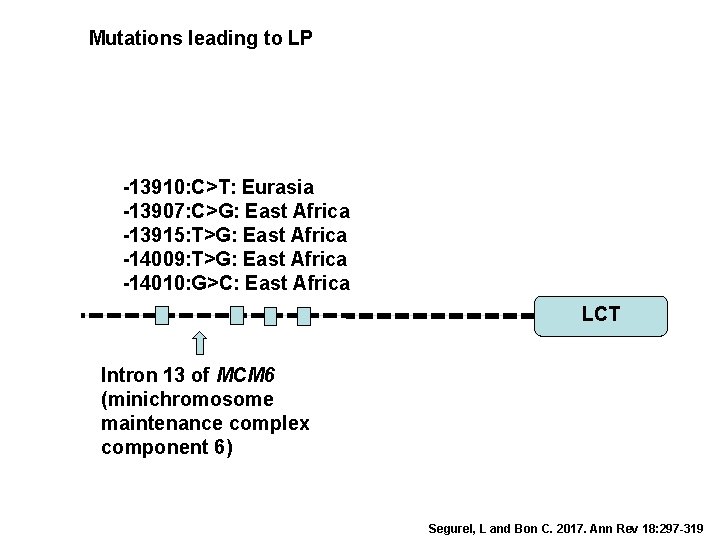
Mutations leading to LP -13910: C>T: Eurasia -13907: C>G: East Africa -13915: T>G: East Africa -14009: T>G: East Africa -14010: G>C: East Africa LCT Intron 13 of MCM 6 (minichromosome maintenance complex component 6) Segurel, L and Bon C. 2017. Ann Rev 18: 297 -319
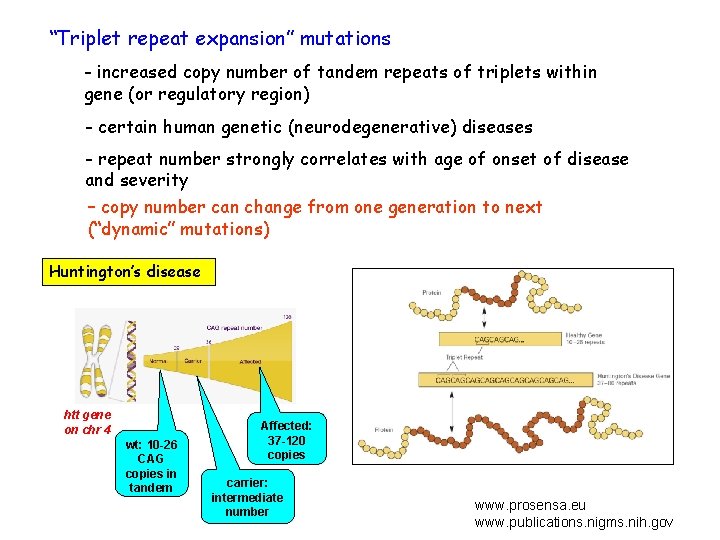
“Triplet repeat expansion” mutations - increased copy number of tandem repeats of triplets within gene (or regulatory region) - certain human genetic (neurodegenerative) diseases - repeat number strongly correlates with age of onset of disease and severity – copy number can change from one generation to next (“dynamic” mutations) Huntington’s disease htt gene on chr 4 wt: 10 -26 CAG copies in tandem Affected: 37 -120 copies carrier: intermediate number www. prosensa. eu www. publications. nigms. nih. gov
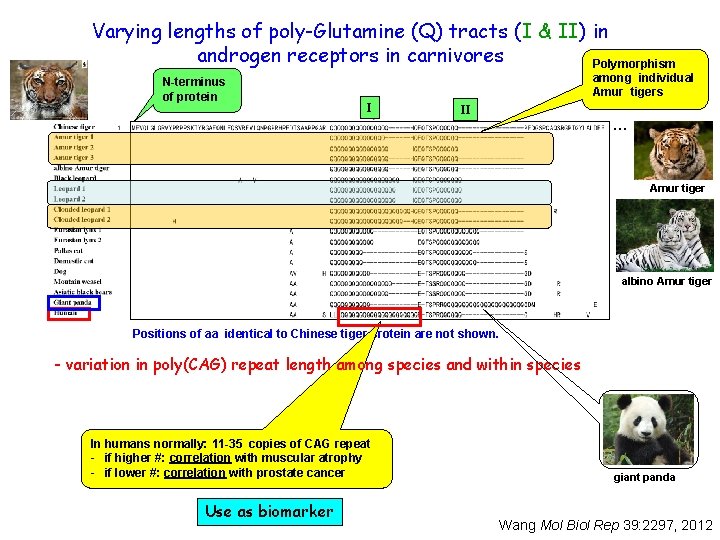
Varying lengths of poly-Glutamine (Q) tracts (I & II) in androgen receptors in carnivores Polymorphism N-terminus of protein among individual Amur tigers I II … Amur tiger albino Amur tiger Positions of aa identical to Chinese tiger protein are not shown. - variation in poly(CAG) repeat length among species and within species In humans normally: 11 -35 copies of CAG repeat - if higher #: correlation with muscular atrophy - if lower #: correlation with prostate cancer Use as biomarker giant panda Wang Mol Biol Rep 39: 2297, 2012
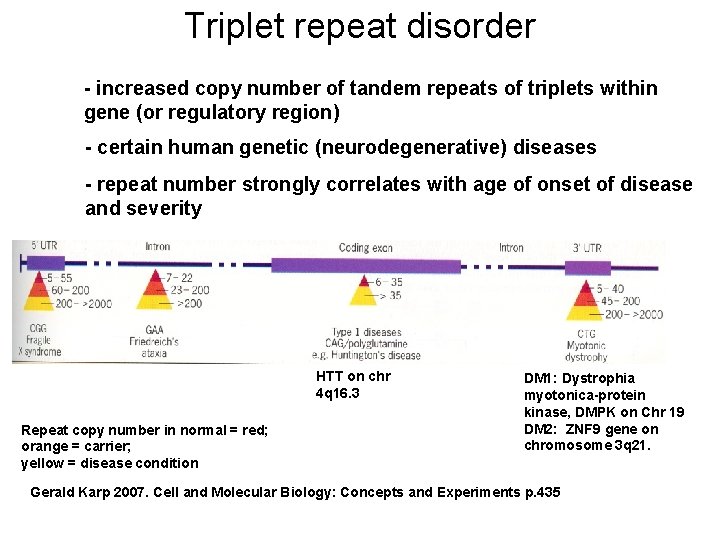
Triplet repeat disorder - increased copy number of tandem repeats of triplets within gene (or regulatory region) - certain human genetic (neurodegenerative) diseases - repeat number strongly correlates with age of onset of disease and severity HTT on chr 4 q 16. 3 Repeat copy number in normal = red; orange = carrier; yellow = disease condition DM 1: Dystrophia myotonica-protein kinase, DMPK on Chr 19 DM 2: ZNF 9 gene on chromosome 3 q 21. Gerald Karp 2007. Cell and Molecular Biology: Concepts and Experiments p. 435
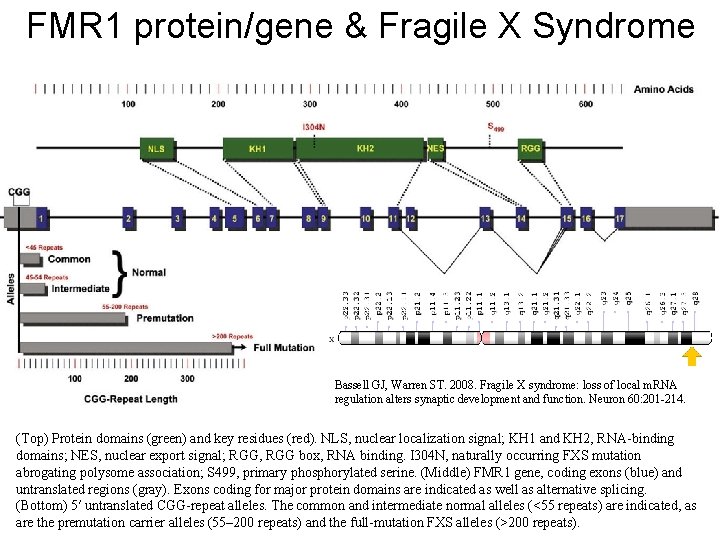
FMR 1 protein/gene & Fragile X Syndrome Bassell GJ, Warren ST. 2008. Fragile X syndrome: loss of local m. RNA regulation alters synaptic development and function. Neuron 60: 201 -214. (Top) Protein domains (green) and key residues (red). NLS, nuclear localization signal; KH 1 and KH 2, RNA-binding domains; NES, nuclear export signal; RGG, RGG box, RNA binding. I 304 N, naturally occurring FXS mutation abrogating polysome association; S 499, primary phosphorylated serine. (Middle) FMR 1 gene, coding exons (blue) and untranslated regions (gray). Exons coding for major protein domains are indicated as well as alternative splicing. (Bottom) 5′ untranslated CGG-repeat alleles. The common and intermediate normal alleles (<55 repeats) are indicated, as are the premutation carrier alleles (55– 200 repeats) and the full-mutation FXS alleles (>200 repeats).
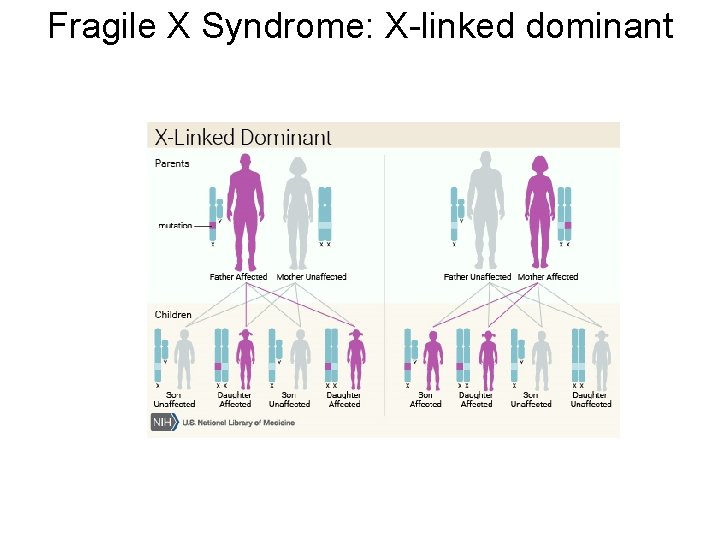
Fragile X Syndrome: X-linked dominant
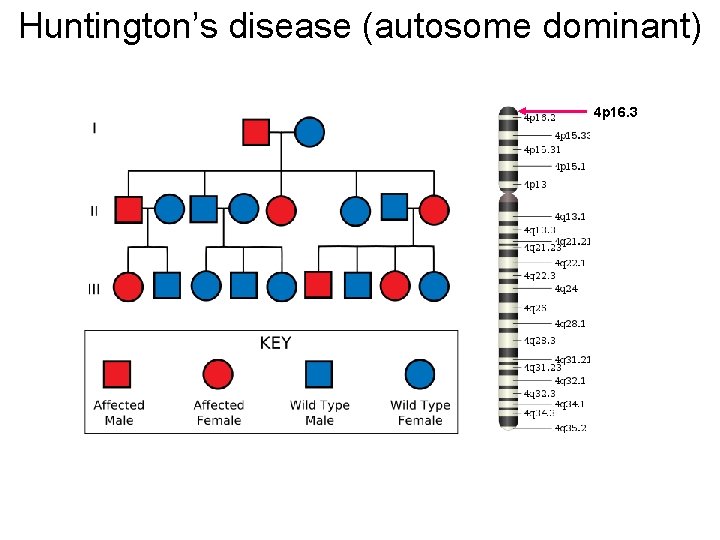
Huntington’s disease (autosome dominant) 4 p 16. 3
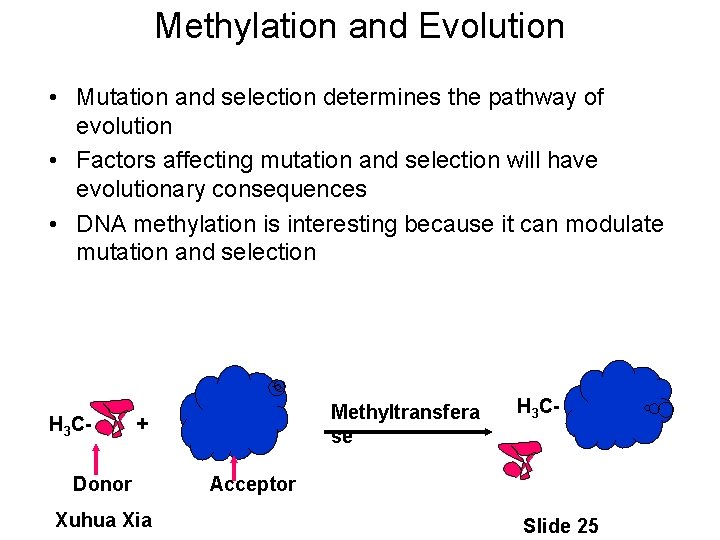
Methylation and Evolution • Mutation and selection determines the pathway of evolution • Factors affecting mutation and selection will have evolutionary consequences • DNA methylation is interesting because it can modulate mutation and selection H 3 C- Methyltransfera se + Donor Xuhua Xia H 3 C- Acceptor Slide 25
Different Methylation Processes • • • Methylation of amino acids in proteins Methylation of nucleotide sequences (DNA and RNA) Differences in methylation sites Differences in functions Illustrations – Methylation and DNA repair in Escherichia coli. – Methylation in the restriction-modification system – Methylation and gene regulation
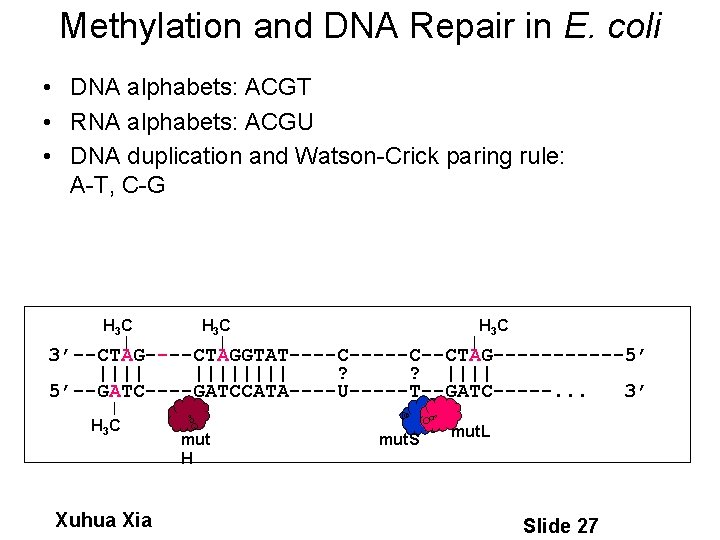
Methylation and DNA Repair in E. coli • DNA alphabets: ACGT • RNA alphabets: ACGU • DNA duplication and Watson-Crick paring rule: A-T, C-G H 3 C H 3 C 3’--CTAG----CTAGGTAT----C--CTAG------5’ |||||||| ? ? |||| 5’--GATC----GATCCATA----U-----T--GATC-----. . . 3’ H 3 C Xuhua Xia mut H mut. S mut. L Slide 27
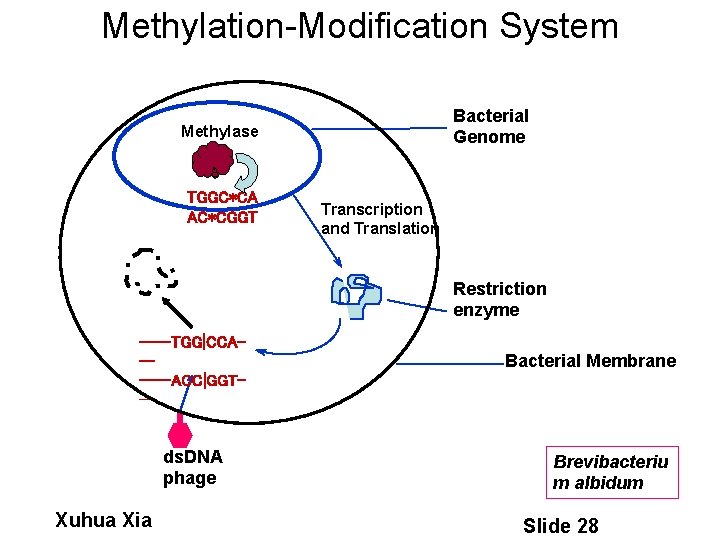
Methylation-Modification System Bacterial Genome Methylase TGGC*CA AC*CGGT Transcription and Translation Restriction enzyme ----TGG|CCA-----ACC|GGT-- ds. DNA phage Xuhua Xia Bacterial Membrane Brevibacteriu m albidum Slide 28
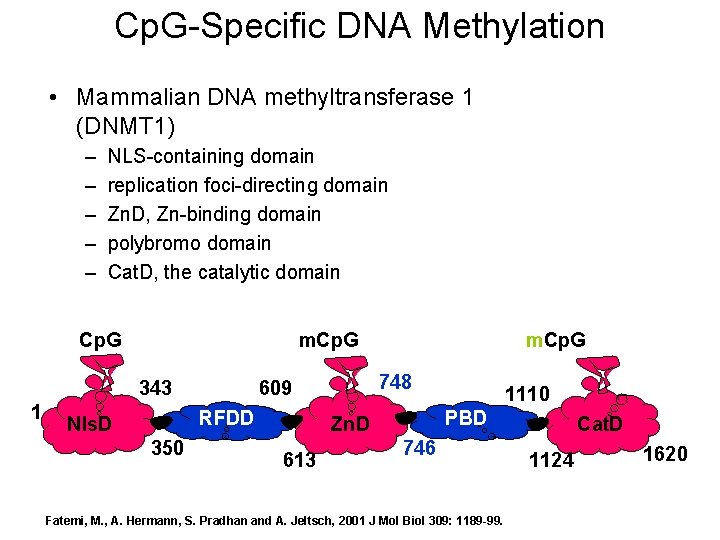
Cp. G-Specific DNA Methylation • Mammalian DNA methyltransferase 1 (DNMT 1) – – – NLS-containing domain replication foci-directing domain Zn. D, Zn-binding domain polybromo domain Cat. D, the catalytic domain Cp. G m. Cp. G 343 1 350 748 609 RFDD Nls. D m. Cp. G PBD Zn. D 613 1110 746 Fatemi, M. , A. Hermann, S. Pradhan and A. Jeltsch, 2001 J Mol Biol 309: 1189 -99. Cat. D 1124 1620
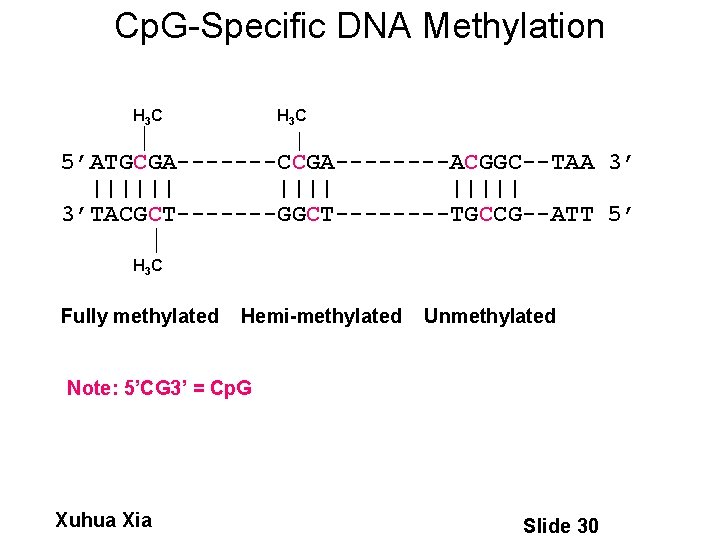
Cp. G-Specific DNA Methylation H 3 C 5’ATGCGA-------CCGA----ACGGC--TAA 3’ |||||| 3’TACGCT-------GGCT----TGCCG--ATT 5’ H 3 C Fully methylated Hemi-methylated Unmethylated Note: 5’CG 3’ = Cp. G Xuhua Xia Slide 30
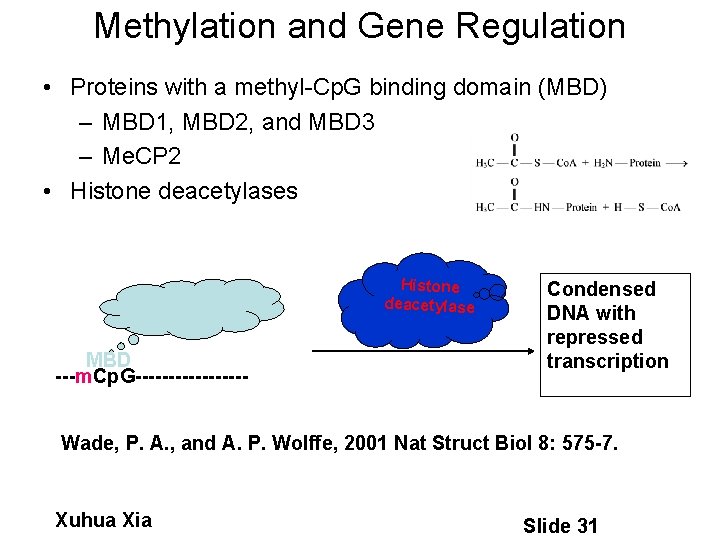
Methylation and Gene Regulation • Proteins with a methyl-Cp. G binding domain (MBD) – MBD 1, MBD 2, and MBD 3 – Me. CP 2 • Histone deacetylases Histone deacetylase MBD ---m. Cp. G--------- Condensed DNA with repressed transcription Wade, P. A. , and A. P. Wolffe, 2001 Nat Struct Biol 8: 575 -7. Xuhua Xia Slide 31
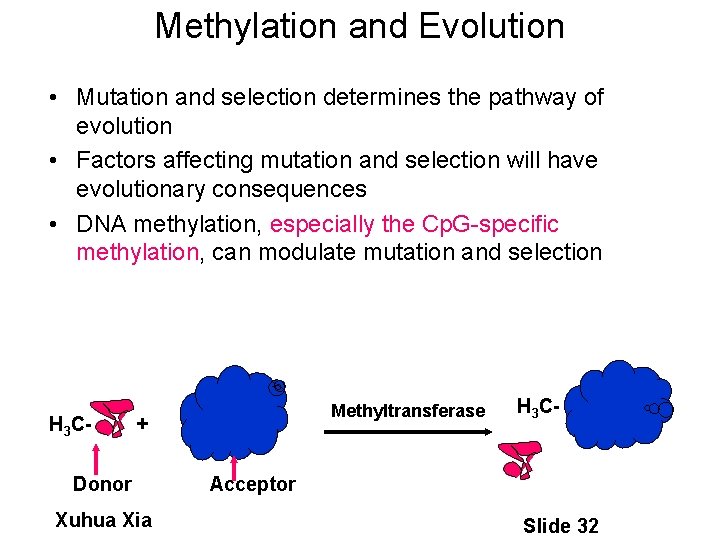
Methylation and Evolution • Mutation and selection determines the pathway of evolution • Factors affecting mutation and selection will have evolutionary consequences • DNA methylation, especially the Cp. G-specific methylation, can modulate mutation and selection H 3 C- Methyltransferase + Donor Xuhua Xia H 3 C- Acceptor Slide 32
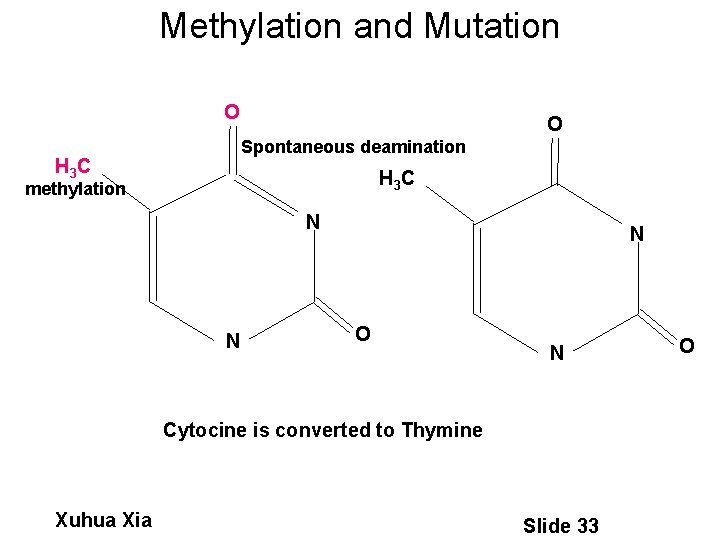
Methylation and Mutation NH O 2 O Spontaneous deamination H 3 C methylation N N N O N Cytocine is converted to Thymine Xuhua Xia Slide 33 O
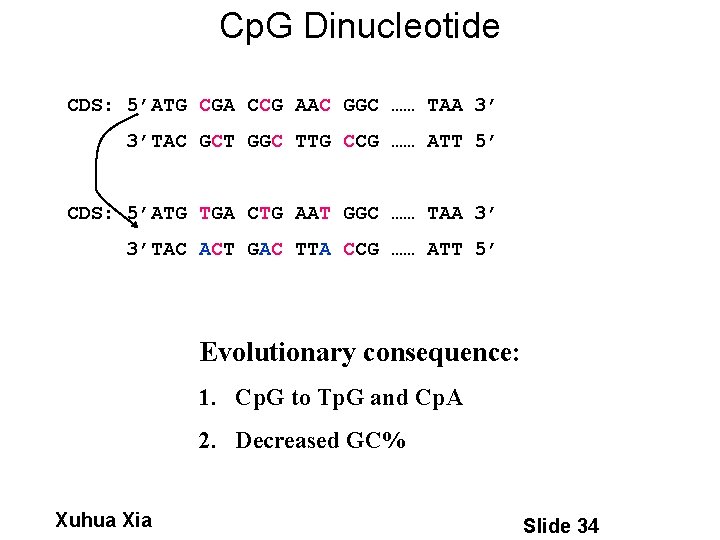
Cp. G Dinucleotide CDS: 5’ATG CGA CCG AAC GGC …… TAA 3’ 3’TAC GCT GGC TTG CCG …… ATT 5’ CDS: 5’ATG TGA CTG AAT GGC …… TAA 3’ 3’TAC ACT GAC TTA CCG …… ATT 5’ Evolutionary consequence: 1. Cp. G to Tp. G and Cp. A 2. Decreased GC% Xuhua Xia Slide 34
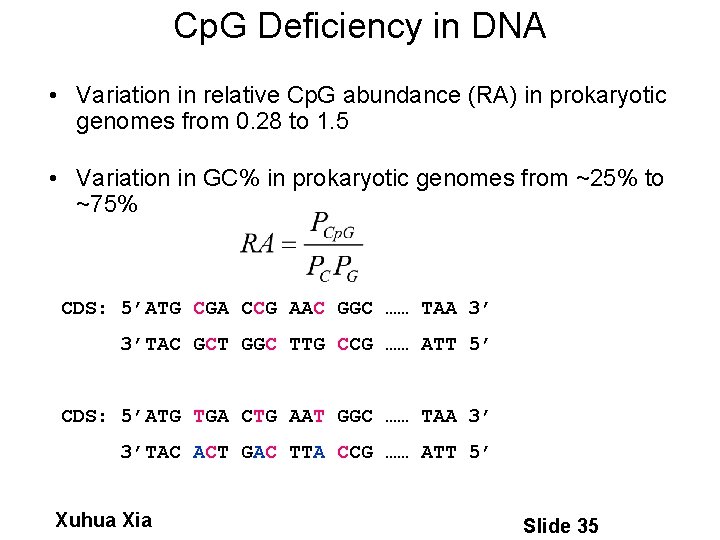
Cp. G Deficiency in DNA • Variation in relative Cp. G abundance (RA) in prokaryotic genomes from 0. 28 to 1. 5 • Variation in GC% in prokaryotic genomes from ~25% to ~75% CDS: 5’ATG CGA CCG AAC GGC …… TAA 3’ 3’TAC GCT GGC TTG CCG …… ATT 5’ CDS: 5’ATG TGA CTG AAT GGC …… TAA 3’ 3’TAC ACT GAC TTA CCG …… ATT 5’ Xuhua Xia Slide 35
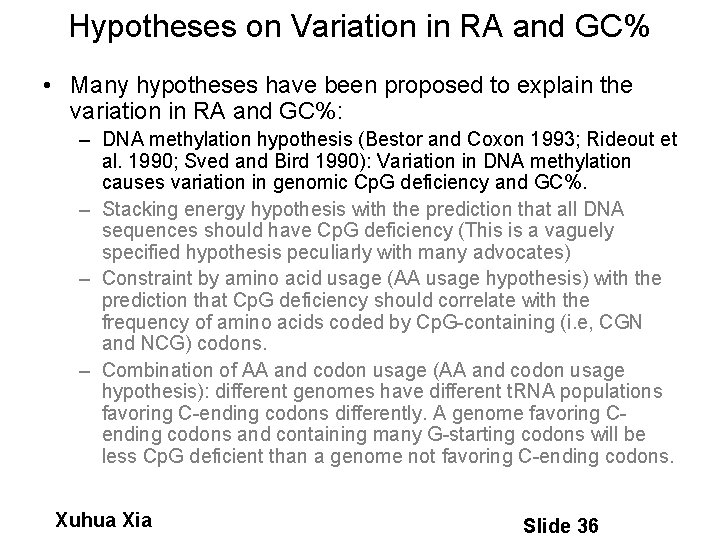
Hypotheses on Variation in RA and GC% • Many hypotheses have been proposed to explain the variation in RA and GC%: – DNA methylation hypothesis (Bestor and Coxon 1993; Rideout et al. 1990; Sved and Bird 1990): Variation in DNA methylation causes variation in genomic Cp. G deficiency and GC%. – Stacking energy hypothesis with the prediction that all DNA sequences should have Cp. G deficiency (This is a vaguely specified hypothesis peculiarly with many advocates) – Constraint by amino acid usage (AA usage hypothesis) with the prediction that Cp. G deficiency should correlate with the frequency of amino acids coded by Cp. G-containing (i. e, CGN and NCG) codons. – Combination of AA and codon usage (AA and codon usage hypothesis): different genomes have different t. RNA populations favoring C-ending codons differently. A genome favoring Cending codons and containing many G-starting codons will be less Cp. G deficient than a genome not favoring C-ending codons. Xuhua Xia Slide 36

Problems • The methylation hypothesis has several major empirical difficulties (e. g. , Cardon et al. 1994), especially in recent years with genome-based analysis (e. g. , Goto et al. 2000): – Great variation in RA between Mycoplasma genitalium and M. pneumoniae. – Extreme Cp. G deficiency in Mycoplasma genitalium Xuhua Xia Slide 37

To Save the Methylation Hypothesis • The methylation hypothesis has to accommodate the arguments above: – How did M. genitalium get strong Cp. G deficiency without methylation? – Why there is so much variation between M. genitalium and M. pneumoniae without involving differential methylation activities? Xuhua Xia Slide 38

He who sees things from the very beginning has the most advantageous view of them. -Aristotle
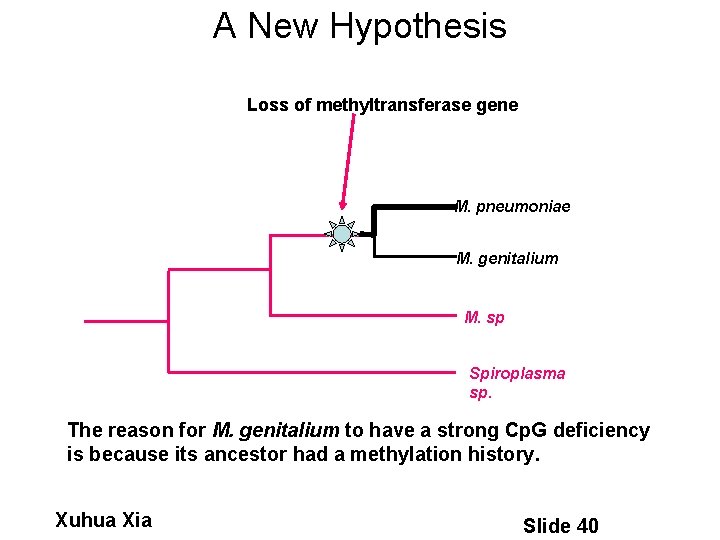
A New Hypothesis Loss of methyltransferase gene M. pneumoniae M. genitalium M. sp Spiroplasma sp. The reason for M. genitalium to have a strong Cp. G deficiency is because its ancestor had a methylation history. Xuhua Xia Slide 40
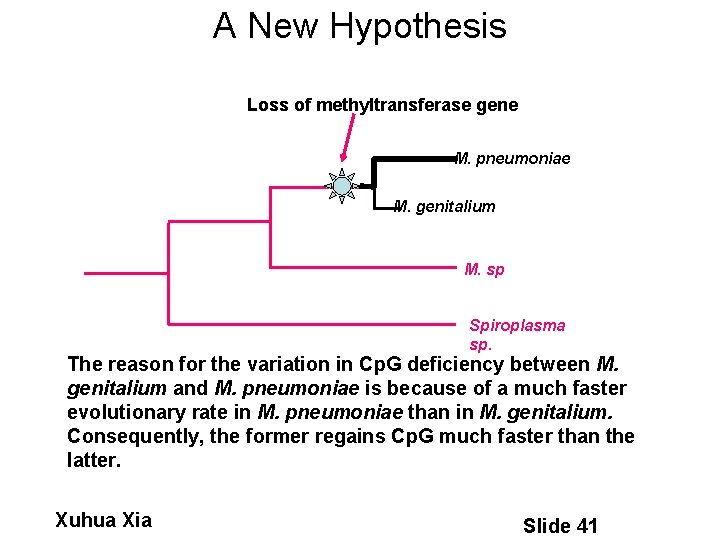
A New Hypothesis Loss of methyltransferase gene M. pneumoniae M. genitalium M. sp Spiroplasma sp. The reason for the variation in Cp. G deficiency between M. genitalium and M. pneumoniae is because of a much faster evolutionary rate in M. pneumoniae than in M. genitalium. Consequently, the former regains Cp. G much faster than the latter. Xuhua Xia Slide 41
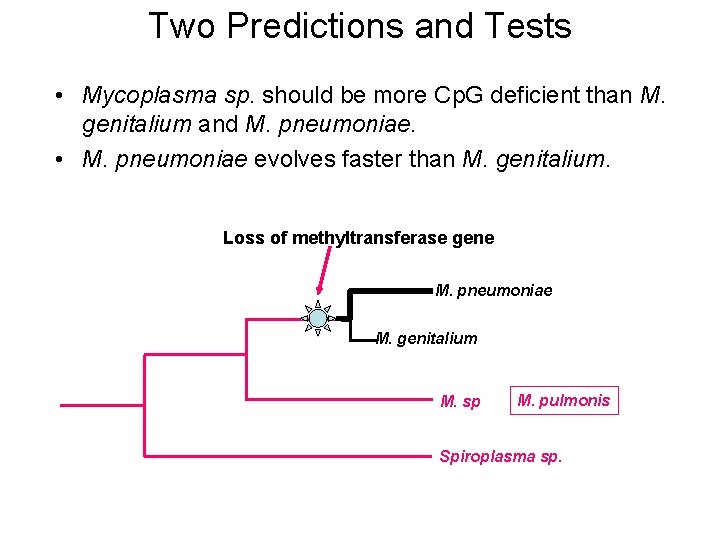
Two Predictions and Tests • Mycoplasma sp. should be more Cp. G deficient than M. genitalium and M. pneumoniae. • M. pneumoniae evolves faster than M. genitalium. Loss of methyltransferase gene M. pneumoniae M. genitalium M. sp M. pulmonis Spiroplasma sp.
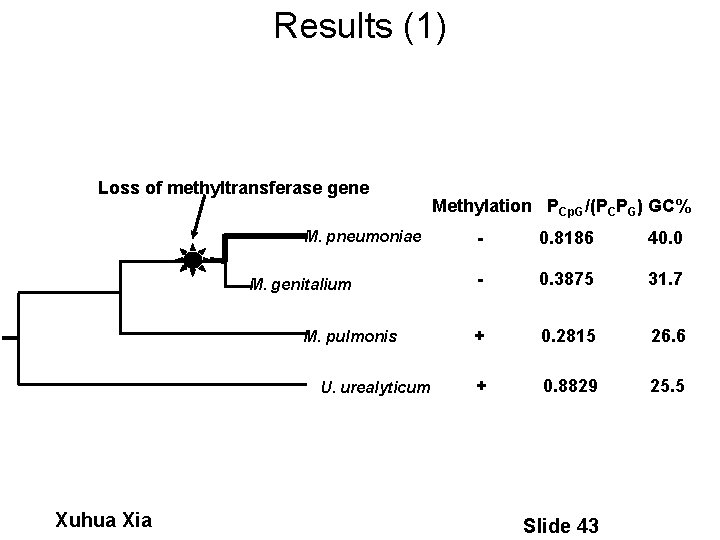
Results (1) Loss of methyltransferase gene M. pneumoniae M. genitalium M. pulmonis U. urealyticum Xuhua Xia Methylation PCp. G/(PCPG) GC% - 0. 8186 40. 0 - 0. 3875 31. 7 + 0. 2815 26. 6 + 0. 8829 25. 5 Slide 43
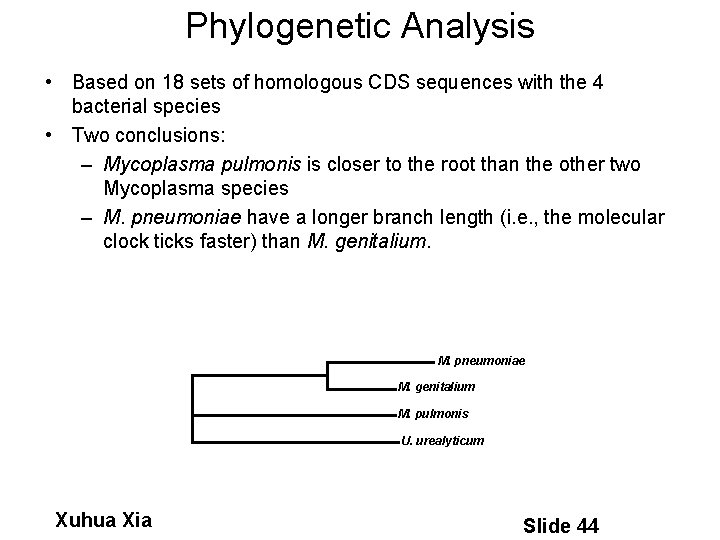
Phylogenetic Analysis • Based on 18 sets of homologous CDS sequences with the 4 bacterial species • Two conclusions: – Mycoplasma pulmonis is closer to the root than the other two Mycoplasma species – M. pneumoniae have a longer branch length (i. e. , the molecular clock ticks faster) than M. genitalium. M. pneumoniae M. genitalium M. pulmonis U. urealyticum Xuhua Xia Slide 44
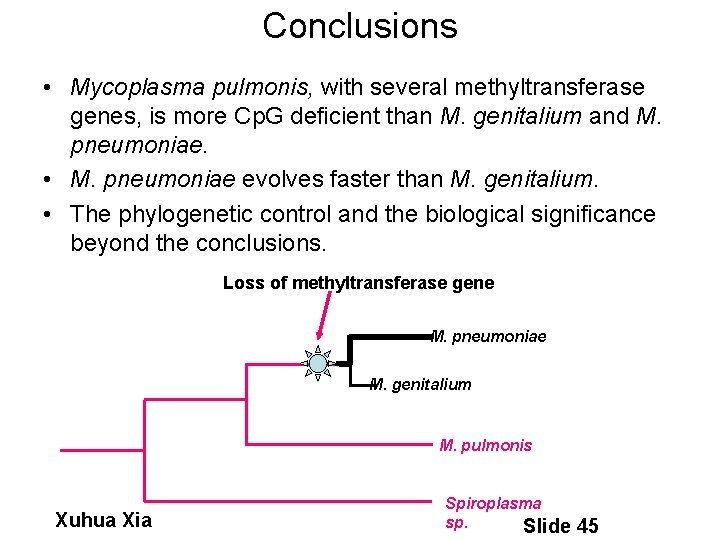
Conclusions • Mycoplasma pulmonis, with several methyltransferase genes, is more Cp. G deficient than M. genitalium and M. pneumoniae. • M. pneumoniae evolves faster than M. genitalium. • The phylogenetic control and the biological significance beyond the conclusions. Loss of methyltransferase gene M. pneumoniae M. genitalium M. pulmonis Xuhua Xia Spiroplasma sp. Slide 45
- Slides: 41