BI 420 Introduction to Bioinformatics Sequence alignment Gabor
BI 420 – Introduction to Bioinformatics Sequence alignment Gabor T. Marth Department of Biology, Boston College marth@bc. edu
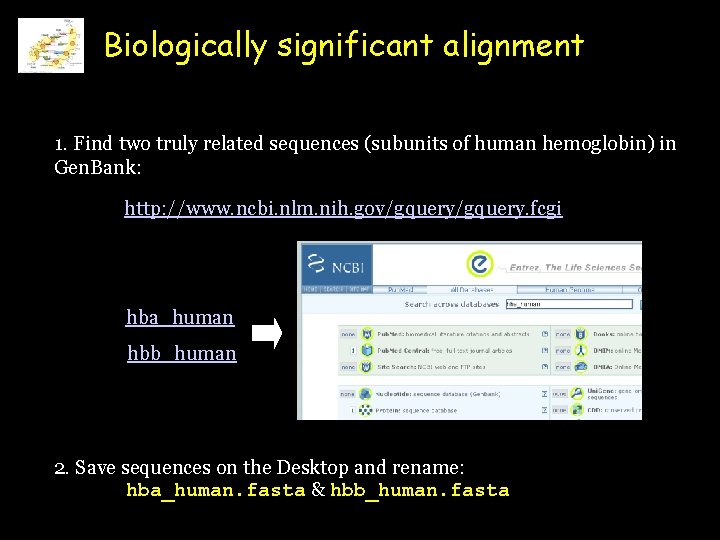
Biologically significant alignment 1. Find two truly related sequences (subunits of human hemoglobin) in Gen. Bank: http: //www. ncbi. nlm. nih. gov/gquery. fcgi hba_human hbb_human 2. Save sequences on the Desktop and rename: hba_human. fasta & hbb_human. fasta
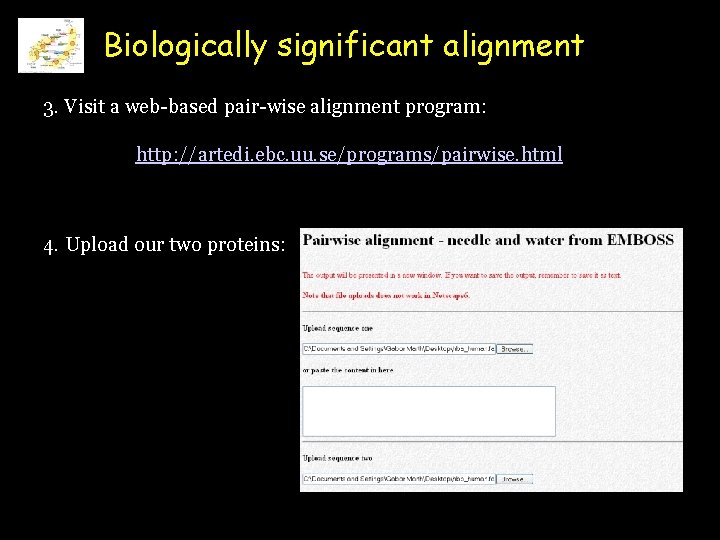
Biologically significant alignment 3. Visit a web-based pair-wise alignment program: http: //artedi. ebc. uu. se/programs/pairwise. html 4. Upload our two proteins:
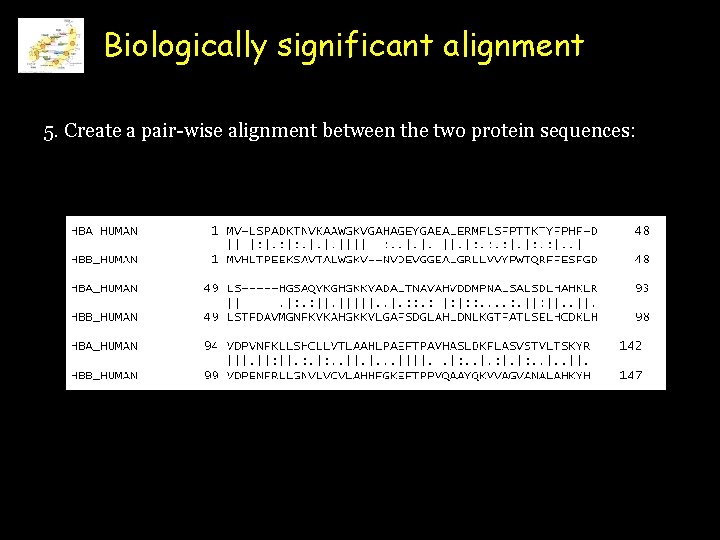
Biologically significant alignment 5. Create a pair-wise alignment between the two protein sequences:

Biologically plausible alignment Retrieve another sequence, leghemoglobin: Leg hemoglobin Create a pair-wise alignment with human hemoglobin A:
Biologically plausible alignment http: //en. wikipedia. org/wiki/Leghemoglobin

Spurious alignment Retrieve the sequence of a human BRCA 1 gene variant, clearly not related to hemoglobin: http: //www. ncbi. nlm. nih. gov/entrez/query. fcgi? db=Protein&cmd=search&term=NP_009225. 1 Make the pair-wise alignment: Examples from: Biological sequence analysis. Durbin, Eddy, Krogh, Mitchison

Alignment types How do we align the words: CRANE and FRAME? CRANE || | FRAME 3 matches, 2 mismatches How do we align words that are different in length? COELACANTH || ||| P-ELICAN-- COELACANTH || ||| -PELICAN-- 5 matches, 2 mismatches, 3 gaps In this case, if we assign +1 points for matches, and -1 for mismatches or gaps, we get 5 x 1 + 1 x (-1) + 3 x (-1) = 0. This is the alignment score. Examples from: BLAST. Korf, Yandell, Bedell

Finding the “best” alignment COELACANTH | ||| PE-LICAN-S=-2 COELACANTH || P-EL-ICANS=-6 COELACANTH || ||| P-ELICAN-S=0 COELACANTH PELICAN-S=-10
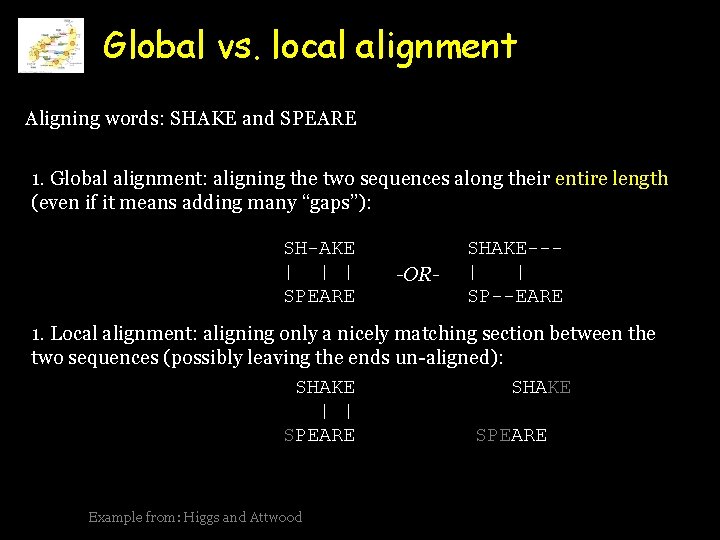
Global vs. local alignment Aligning words: SHAKE and SPEARE 1. Global alignment: aligning the two sequences along their entire length (even if it means adding many “gaps”): SH-AKE | | | SPEARE -OR- SHAKE--| | SP--EARE 1. Local alignment: aligning only a nicely matching section between the two sequences (possibly leaving the ends un-aligned): SHAKE | | SPEARE Example from: Higgs and Attwood

Global alignment – Needleman-Wunsch + gap score g = -6 Pair-wise amino-acid scores S(ai, bi) (PAM 250 scoring scheme) plus gap score g. Example from: Higgs and Attwood
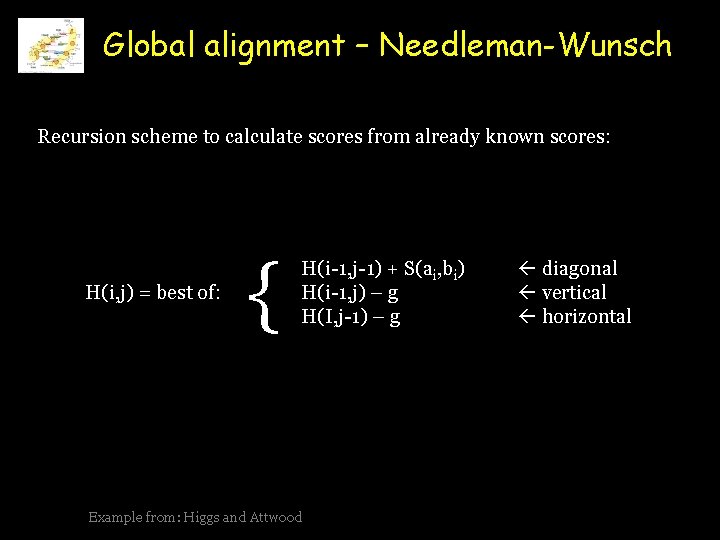
Global alignment – Needleman-Wunsch Recursion scheme to calculate scores from already known scores: H(i, j) = best of: { H(i-1, j-1) + S(ai, bi) H(i-1, j) – g H(I, j-1) – g Example from: Higgs and Attwood diagonal vertical horizontal

Global alignment – Needleman-Wunsch Initialization (filling the top row and left column from gap scores): Align the two sequences: AAGATTCAC and CCGCTCAA Example from: Higgs and Attwood
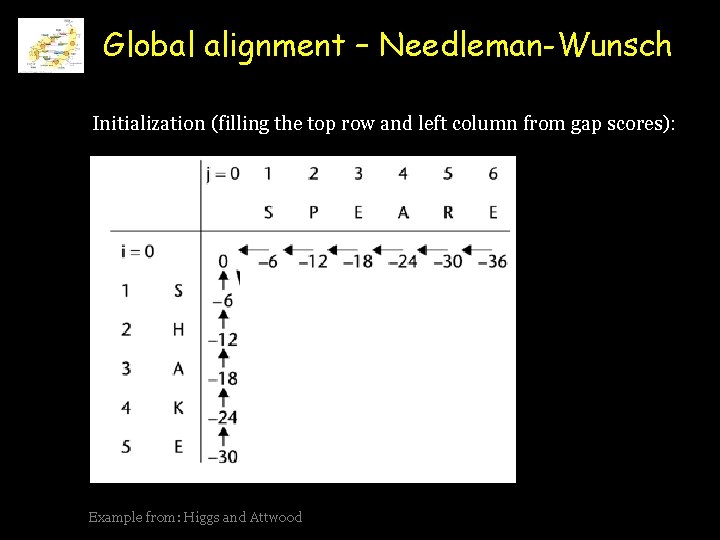
Global alignment – Needleman-Wunsch Initialization (filling the top row and left column from gap scores): Example from: Higgs and Attwood
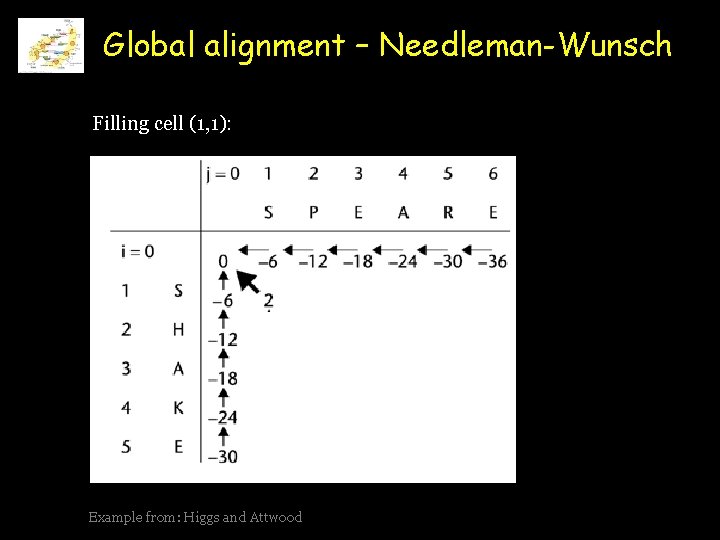
Global alignment – Needleman-Wunsch Filling cell (1, 1): Example from: Higgs and Attwood
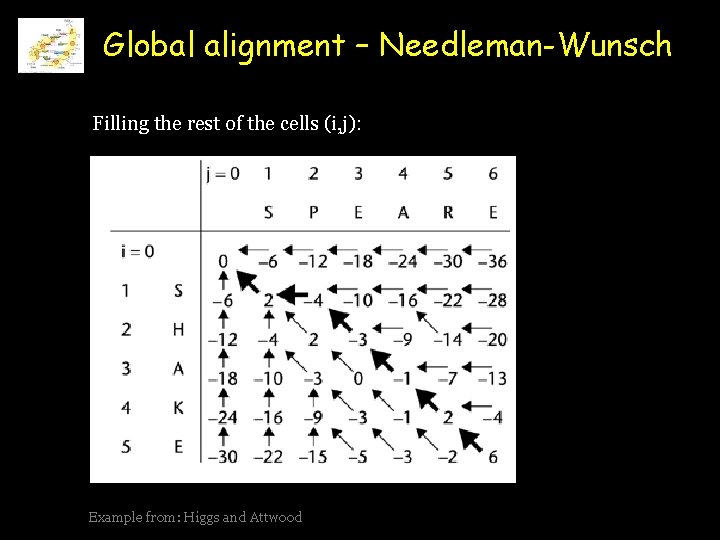
Global alignment – Needleman-Wunsch Filling the rest of the cells (i, j): Example from: Higgs and Attwood

Global alignment – Needleman-Wunsch Tracing back to read out the alignment: Best global alignment: S-HAKE SPEARE Example from: Higgs and Attwood
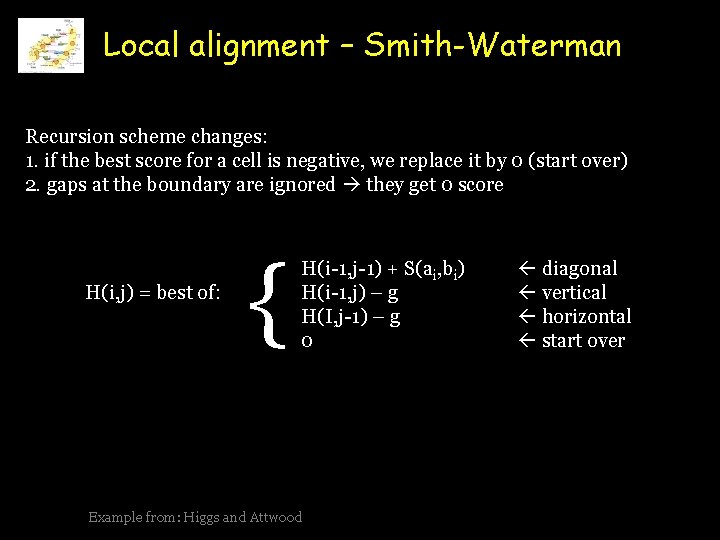
Local alignment – Smith-Waterman Recursion scheme changes: 1. if the best score for a cell is negative, we replace it by 0 (start over) 2. gaps at the boundary are ignored they get 0 score H(i, j) = best of: { H(i-1, j-1) + S(ai, bi) H(i-1, j) – g H(I, j-1) – g 0 Example from: Higgs and Attwood diagonal vertical horizontal start over

Local alignment – Smith-Waterman Initialization Example from: Higgs and Attwood

Local alignment – Smith-Waterman Align the two sequences: AAGATTCAC and CCGCTCAA Initialization Example from: Higgs and Attwood
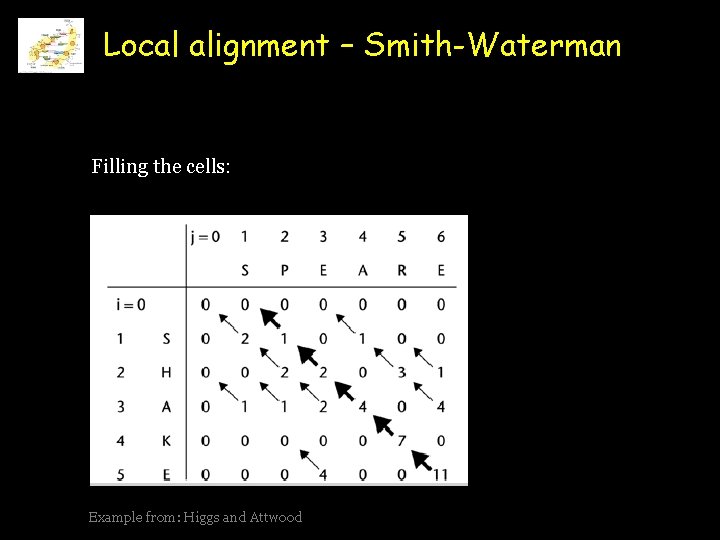
Local alignment – Smith-Waterman Filling the cells: Example from: Higgs and Attwood

Local alignment – Smith-Waterman Trace-back: Best local alignment: SHAKE SPEARE Example from: Higgs and Attwood
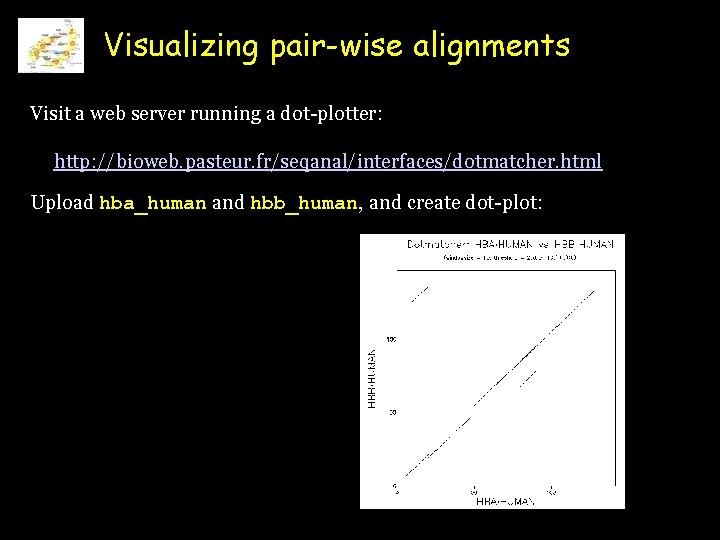
Visualizing pair-wise alignments Visit a web server running a dot-plotter: http: //bioweb. pasteur. fr/seqanal/interfaces/dotmatcher. html Upload hba_human and hbb_human, and create dot-plot:

Scoring schemes Match-mismatch-gap penalties: e. g. Match = 1 Mismatch = -5 Gap = -10 Scoring matrices
Multiple alignments Fetch HXK (hexokinase) sequences from NCBI; save as hxk. fasta on the Desktop
Multiple alignments Visit a web-hosted clustal. W site (e. g. : http: //artedi. ebc. uu. se/programs/clustalw. html) and upload the HXK sequences

Multiple alignments The multiple alignment of 24 hexokinese protein sequences from various species
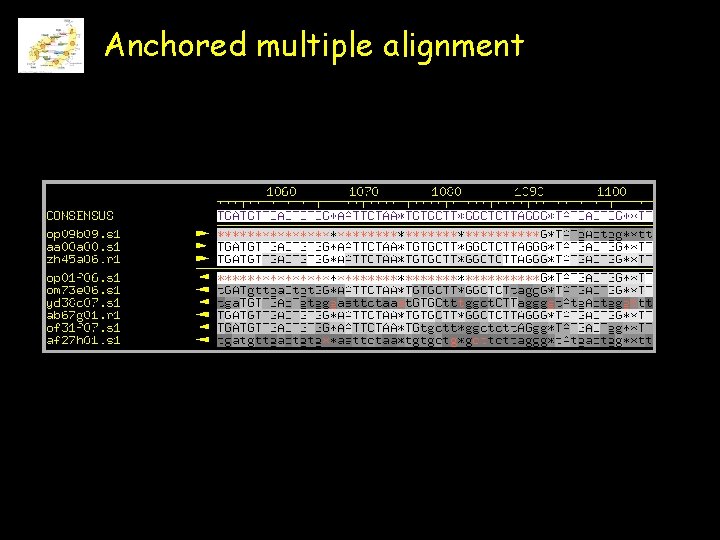
Anchored multiple alignment
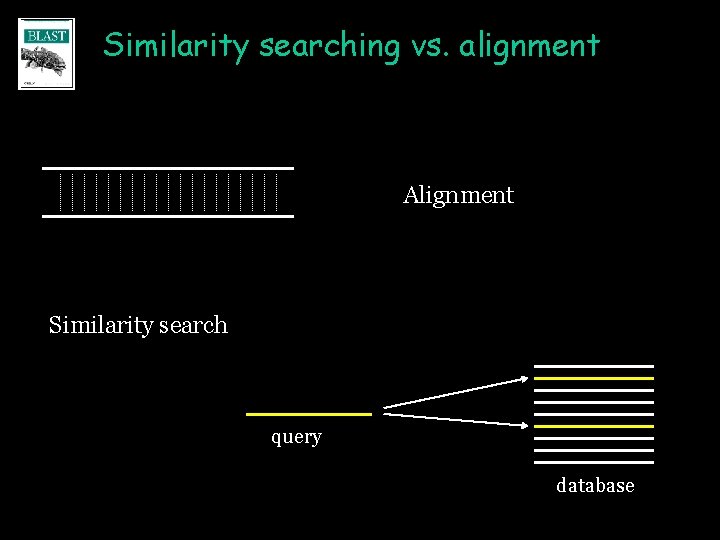
Similarity searching vs. alignment Alignment Similarity search query database
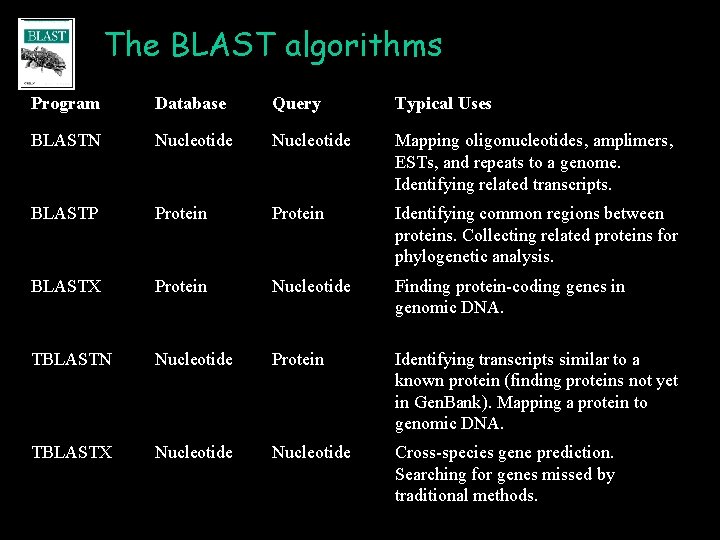
The BLAST algorithms Program Database Query Typical Uses BLASTN Nucleotide Mapping oligonucleotides, amplimers, ESTs, and repeats to a genome. Identifying related transcripts. BLASTP Protein Identifying common regions between proteins. Collecting related proteins for phylogenetic analysis. BLASTX Protein Nucleotide Finding protein-coding genes in genomic DNA. TBLASTN Nucleotide Protein Identifying transcripts similar to a known protein (finding proteins not yet in Gen. Bank). Mapping a protein to genomic DNA. TBLASTX Nucleotide Cross-species gene prediction. Searching for genes missed by traditional methods.

BLAST report http: //www. ncbi. nlm. nih. gov/BLAST/ gi|7428631

BLAST report

The BLAST algorithm Sequence alignment takes place in a 2 -dimensional space where diagonal lines represent regions of similarity. Gaps in an alignment appear as broken diagonals. The search space is sometimes considered as 2 sequences and somtimes as query x database. • Global alignment vs. local alignment – BLAST is local • Maximum scoring pair (MSP) vs. High-scoring pair (HSP) – BLAST finds HSPs (usually the MSP too) • Gapped vs. ungapped – BLAST can do both

The BLAST algorithm • Speed gained by minimizing search space • Alignments require word hits • Neighborhood words T=12 • W and T modulate speed and sensitivity BLOSUM 62 neighborhood of RGD 17 KGD 14 QGD 13 RGE 13 EGD 12 HGD 12 NGD 12 RGN 12 AGD 11 MGD 11 RAD 11 RGQ 11 RGS 11 RND 11 RSD 11 SGD 11 TGD 11
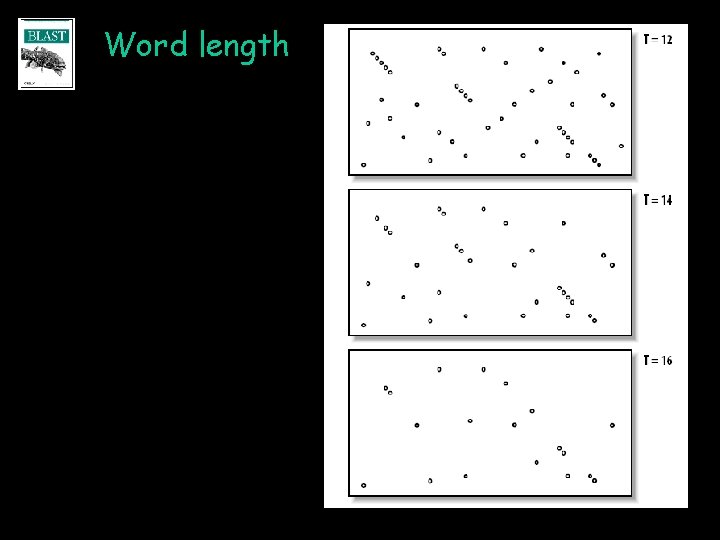
Word length

2 -hit seeding • Alignments tend to have multiple word hits. • Isolated word hits are frequently false leads. • Most alignments have large ungapped regions. • Requiring 2 word hits on the same diagonal (of 40 aa for example), greatly increases speed at a slight cost in sensitivity.

Extension of the seed alignments • Alignments are extended from seeds in each direction. • Extension is terminated when the maximum score drops below X. Text example match +1 mismatch -1 no gaps The quick brown fox jumps over the lazy dog. The quiet brown cat purrs when she sees him.
![BLAST statistics >gi|23098447|ref|NP_691913. 1| (NC_004193) 3 -oxoacyl-(acyl carrier protein) reductase [Oceanobacillus iheyensis] Length = BLAST statistics >gi|23098447|ref|NP_691913. 1| (NC_004193) 3 -oxoacyl-(acyl carrier protein) reductase [Oceanobacillus iheyensis] Length =](http://slidetodoc.com/presentation_image/41b5e66b014f16f685d0f420911bd059/image-38.jpg)
BLAST statistics >gi|23098447|ref|NP_691913. 1| (NC_004193) 3 -oxoacyl-(acyl carrier protein) reductase [Oceanobacillus iheyensis] Length = 253 Score = 38. 9 bits (89), Expect = 3 e-05 Identities = 17/40 (42%), Positives = 26/40 (64%) Frame = -1 Query: 4146 VTGAGHGLGRAISLELAKKGCHIAVVDINVSGAEDTVKQI 4027 VTGA G+G+AI+ A +G + V D+N GA+ V++I Sbjct: 10 VTGAASGMGKAIATLYASEGAKVIVADLNEEGAQSVVEEI 49 How significant is this similarity?

Scoring the alignment Query: 4146 VTGAGHGLGRAISLELAKKGCHIAVVDINVSGAEDTVKQI 4027 VTGA G+G+AI+ A +G + V D+N GA+ V++I Sbjct: 10 VTGAASGMGKAIATLYASEGAKVIVADLNEEGAQSVVEEI 49 4 -1 4 S (score)
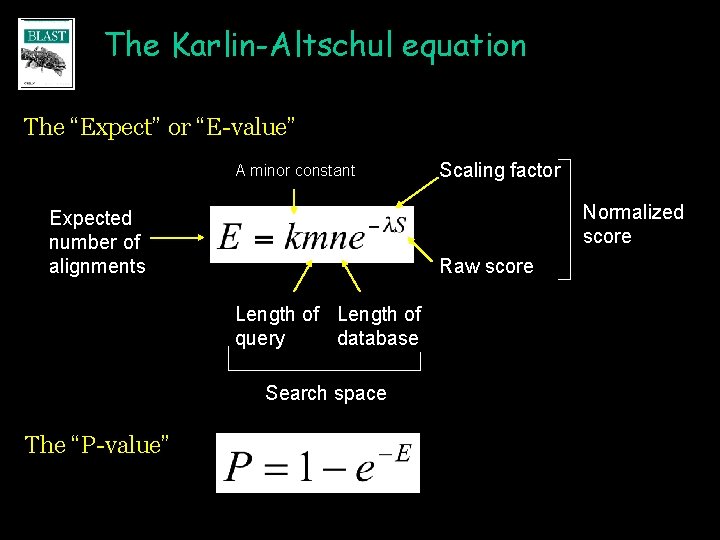
The Karlin-Altschul equation The “Expect” or “E-value” A minor constant Normalized score Expected number of alignments Raw score Length of query database Search space The “P-value” Scaling factor
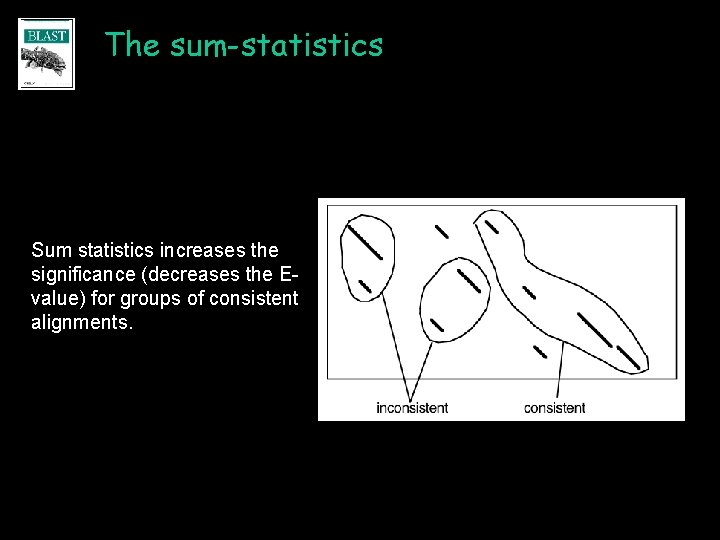
The sum-statistics Sum statistics increases the significance (decreases the Evalue) for groups of consistent alignments.
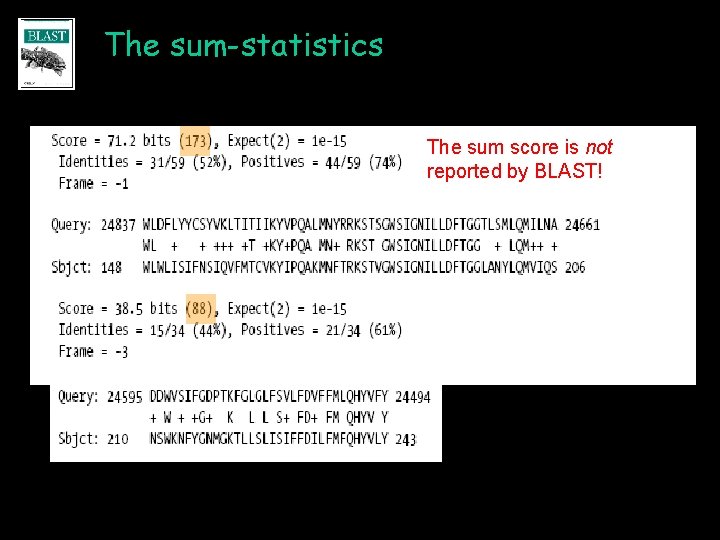
The sum-statistics The sum score is not reported by BLAST!
- Slides: 42