Best Practices for the Use of Clinical Databases

Best Practices for the Use of Clinical Databases for Variant Classification and Interpretation in Clinical Oncology Karla R. Bowles, Ph. D, FACMG Senior Laboratory Director Myriad Genetic Laboratories, Inc. Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com

Disclosures I am employed by Myriad Genetic Laboratories, Inc. and receive salary and stock options as compensation. Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com

Variant Databases and Associated Tools Support Variant Classification and Reclassification Initial Variant Observation Variant Classification (5 -Tier System) • Pathogenic • Benign • VUS • Likely Pathogenic • Likely Benign Variant Reclassification • Gather sufficient data • Update variant-specific clinical interpretation Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com
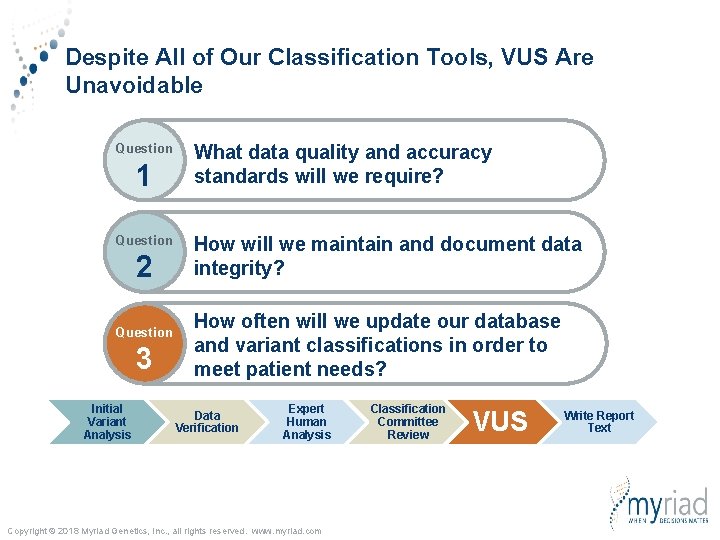
Key Questions When Developing a Clinical Database Question 1 Question 2 Question 3 What data quality and accuracy standards will we require? How will we maintain and document data integrity? How often will we update our database and variant classifications in order to meet patient needs? Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com

Key Questions When Developing a Clinical Database Question 1 Question 2 Question 3 What data quality and accuracy standards will we require? How will we maintain and document data integrity? How often will we update our database and variant classifications in order to meet patient needs? Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com
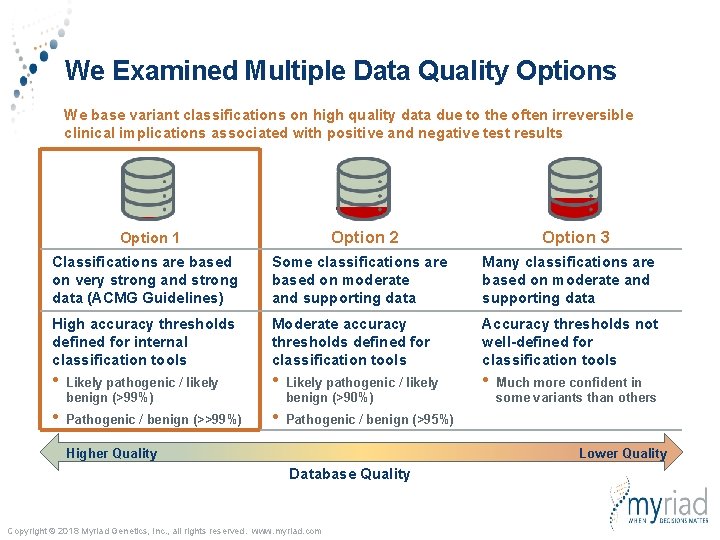
We Examined Multiple Data Quality Options We base variant classifications on high quality data due to the often irreversible clinical implications associated with positive and negative test results Option 2 Option 1 Option 3 Classifications are based on very strong and strong data (ACMG Guidelines) Some classifications are based on moderate and supporting data Many classifications are based on moderate and supporting data High accuracy thresholds defined for internal classification tools Moderate accuracy thresholds defined for classification tools Accuracy thresholds not well-defined for classification tools • Likely pathogenic / likely benign (>99%) • Likely pathogenic / likely benign (>90%) • • Pathogenic / benign (>>99%) • Pathogenic / benign (>95%) Higher Quality Much more confident in some variants than others Lower Quality Database Quality Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com

We Establish Classification Confidence Thresholds Before Data is Used We Estimate Accuracy for Each of Our Classification Tools • • We evaluate tools independently using large numbers of control variants with well-established classifications Tools are independently assessed Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com

We Establish Classification Confidence Thresholds Before Data is Used We Estimate Accuracy for Each of Our Classification Tools • • We Estimate Tool • Accuracy for Each Gene • We evaluate tools independently using large numbers of control variants with well-established classifications Tools are independently assessed Some tools work better for some genes than others It is not always safe to assume that accuracy is uniform for all genes Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com

We Establish Classification Confidence Thresholds Before Data is Used We Estimate Accuracy for Each of Our Classification Tools • • We Estimate Tool • Accuracy for Each Gene • • Tool Accuracy Estimates are Based on Clinical Effect(s) • • We evaluate tools independently using large numbers of control variants with well-established classifications Tools are independently assessed Some tools work better for some genes than others It is not always safe to assume that accuracy is uniform for all genes Variants may negatively impact protein production and/or function without resulting in increased cancer risk There must be a direct connection between the classification tool and estimation of cancer risk We often exclude tools based on lower model organisms (e. g. yeast) Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com

We Establish Classification Confidence Thresholds Before Data is Used We Estimate Accuracy for Each of Our Classification Tools • • We Estimate Tool • Accuracy for Each Gene • • Tool Accuracy Estimates are Based on Clinical Effect(s) • • We Use Non-biased Tools Whenever Possible • • We evaluate tools independently using large numbers of control variants with well-established classifications Tools are independently assessed Some tools work better for some genes than others It is not always safe to assume that accuracy is uniform for all genes Variants may negatively impact protein production and/or function without resulting in increased cancer risk There must be a direct connection between the classification tool and estimation of cancer risk We often exclude tools based on lower model organisms (e. g. yeast) High quality statistical tools have quantifiable high accuracy Tools requiring significant human interpretation have a greater chance of error Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com
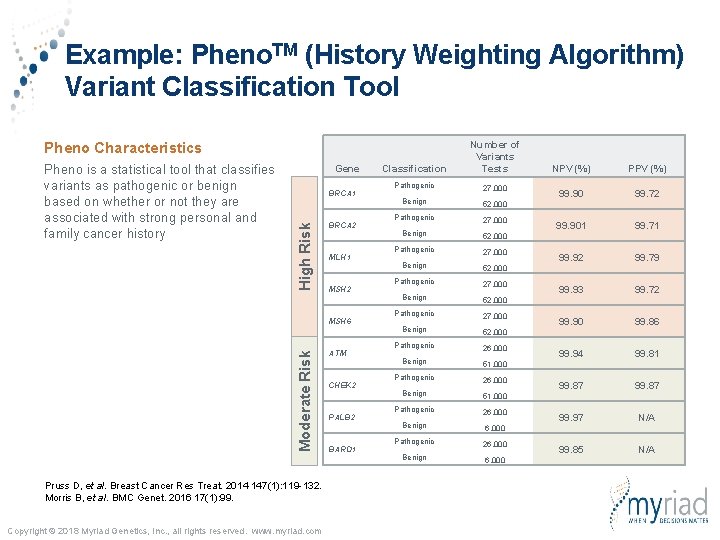
Example: Pheno. TM (History Weighting Algorithm) Variant Classification Tool Classification Number of Variants Tests Pathogenic 27, 000 Benign 52, 000 Pathogenic 27, 000 Benign 52, 000 Pathogenic 26, 000 Benign 51, 000 Pathogenic 26, 000 Benign 6, 000 Pheno Characteristics Gene BRCA 1 High Risk Pheno is a statistical tool that classifies variants as pathogenic or benign based on whether or not they are associated with strong personal and family cancer history BRCA 2 MLH 1 MSH 2 Moderate Risk MSH 6 Pruss D, et al. Breast Cancer Res Treat. 2014 147(1): 119 -132. Morris B, et al. BMC Genet. 2016 17(1): 99. Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com ATM CHEK 2 PALB 2 BARD 1 NPV (%) PPV (%) 99. 90 99. 72 99. 901 99. 71 99. 92 99. 79 99. 93 99. 72 99. 90 99. 86 99. 94 99. 81 99. 87 99. 97 N/A 99. 85 N/A
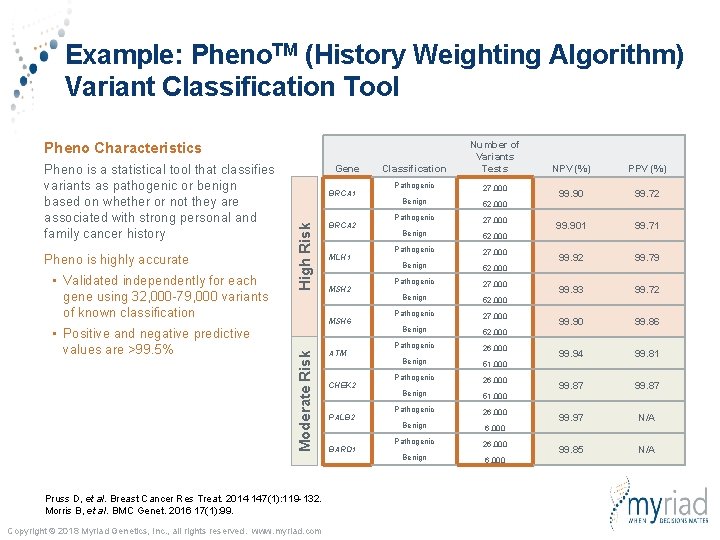
Example: Pheno. TM (History Weighting Algorithm) Variant Classification Tool Classification Number of Variants Tests Pathogenic 27, 000 Benign 52, 000 Pathogenic 27, 000 Benign 52, 000 Pathogenic 26, 000 Benign 51, 000 Pathogenic 26, 000 Benign 6, 000 Pheno Characteristics • Validated independently for each gene using 32, 000 -79, 000 variants of known classification • Positive and negative predictive values are >99. 5% BRCA 1 High Risk Pheno is highly accurate Gene BRCA 2 MLH 1 MSH 2 MSH 6 Moderate Risk Pheno is a statistical tool that classifies variants as pathogenic or benign based on whether or not they are associated with strong personal and family cancer history Pruss D, et al. Breast Cancer Res Treat. 2014 147(1): 119 -132. Morris B, et al. BMC Genet. 2016 17(1): 99. Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com ATM CHEK 2 PALB 2 BARD 1 NPV (%) PPV (%) 99. 90 99. 72 99. 901 99. 71 99. 92 99. 79 99. 93 99. 72 99. 90 99. 86 99. 94 99. 81 99. 87 99. 97 N/A 99. 85 N/A
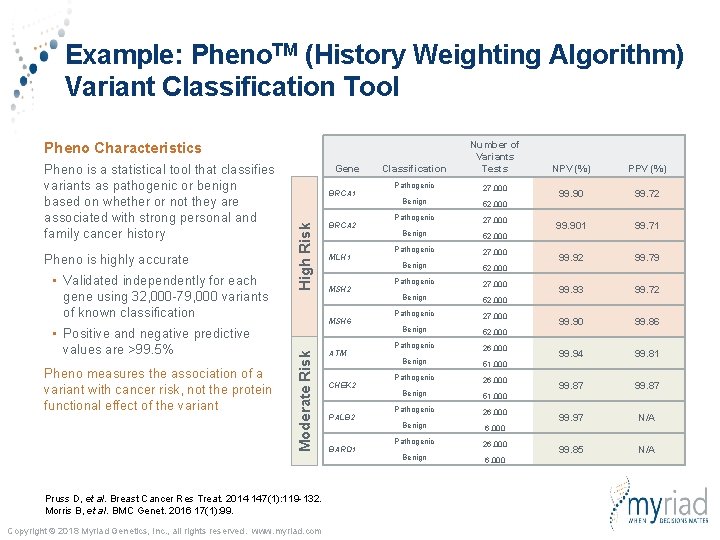
Example: Pheno. TM (History Weighting Algorithm) Variant Classification Tool Classification Number of Variants Tests Pathogenic 27, 000 Benign 52, 000 Pathogenic 27, 000 Benign 52, 000 Pathogenic 26, 000 Benign 51, 000 Pathogenic 26, 000 Benign 6, 000 Pheno Characteristics • Validated independently for each gene using 32, 000 -79, 000 variants of known classification • Positive and negative predictive values are >99. 5% Pheno measures the association of a variant with cancer risk, not the protein functional effect of the variant BRCA 1 High Risk Pheno is highly accurate Gene BRCA 2 MLH 1 MSH 2 MSH 6 Moderate Risk Pheno is a statistical tool that classifies variants as pathogenic or benign based on whether or not they are associated with strong personal and family cancer history Pruss D, et al. Breast Cancer Res Treat. 2014 147(1): 119 -132. Morris B, et al. BMC Genet. 2016 17(1): 99. Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com ATM CHEK 2 PALB 2 BARD 1 NPV (%) PPV (%) 99. 90 99. 72 99. 901 99. 71 99. 92 99. 79 99. 93 99. 72 99. 90 99. 86 99. 94 99. 81 99. 87 99. 97 N/A 99. 85 N/A
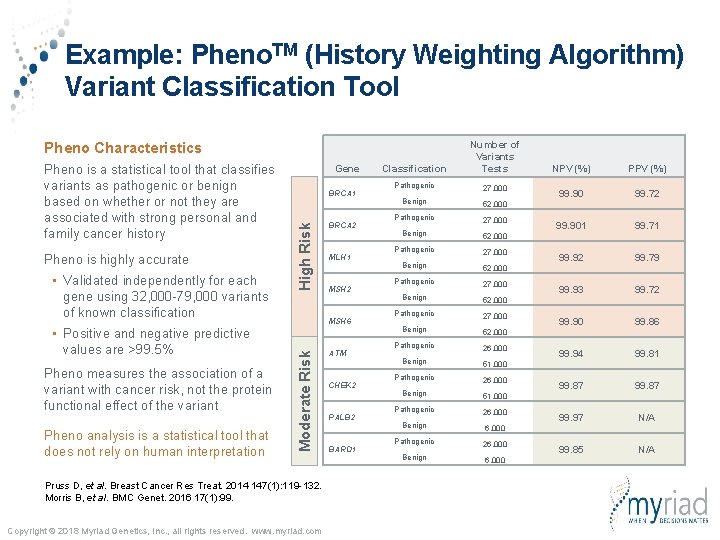
Example: Pheno. TM (History Weighting Algorithm) Variant Classification Tool Classification Number of Variants Tests Pathogenic 27, 000 Benign 52, 000 Pathogenic 27, 000 Benign 52, 000 Pathogenic 26, 000 Benign 51, 000 Pathogenic 26, 000 Benign 6, 000 Pheno Characteristics • Validated independently for each gene using 32, 000 -79, 000 variants of known classification • Positive and negative predictive values are >99. 5% Pheno measures the association of a variant with cancer risk, not the protein functional effect of the variant Pheno analysis is a statistical tool that does not rely on human interpretation BRCA 1 High Risk Pheno is highly accurate Gene BRCA 2 MLH 1 MSH 2 MSH 6 Moderate Risk Pheno is a statistical tool that classifies variants as pathogenic or benign based on whether or not they are associated with strong personal and family cancer history Pruss D, et al. Breast Cancer Res Treat. 2014 147(1): 119 -132. Morris B, et al. BMC Genet. 2016 17(1): 99. Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com ATM CHEK 2 PALB 2 BARD 1 NPV (%) PPV (%) 99. 90 99. 72 99. 901 99. 71 99. 92 99. 79 99. 93 99. 72 99. 90 99. 86 99. 94 99. 81 99. 87 99. 97 N/A 99. 85 N/A
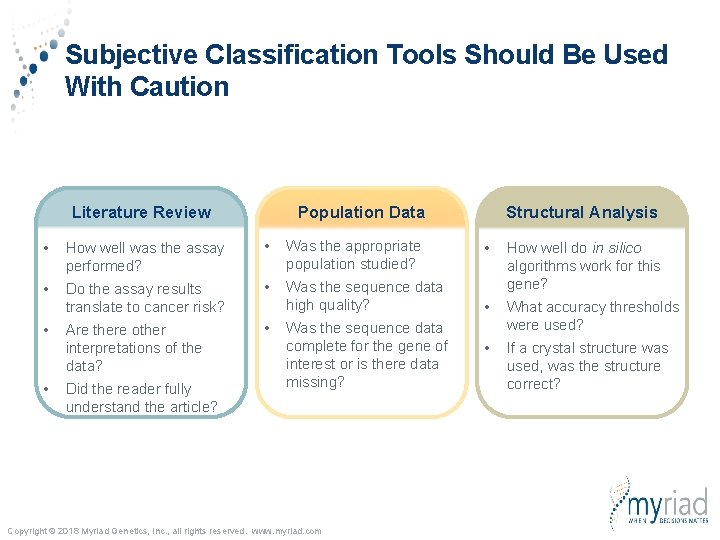
Subjective Classification Tools Should Be Used With Caution Literature Review Population Data • How well was the assay performed? • Was the appropriate population studied? • Do the assay results translate to cancer risk? • Was the sequence data high quality? Are there other interpretations of the data? • • • Did the reader fully understand the article? Was the sequence data complete for the gene of interest or is there data missing? Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com Structural Analysis • How well do in silico algorithms work for this gene? • What accuracy thresholds were used? • If a crystal structure was used, was the structure correct?

Key Questions When Developing a Clinical Database Question 1 Question 2 Question 3 What data quality and accuracy standards will we require? How will we maintain and document data integrity? How often will we update our database and variant classifications in order to meet patient needs? Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com
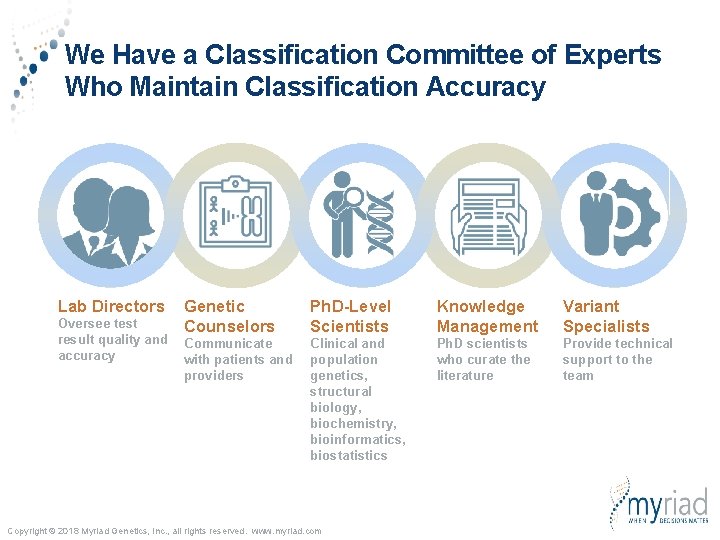
We Have a Classification Committee of Experts Who Maintain Classification Accuracy Lab Directors Oversee test result quality and accuracy Genetic Counselors Ph. D-Level Scientists Knowledge Management Variant Specialists Communicate with patients and providers Clinical and population genetics, structural biology, biochemistry, bioinformatics, biostatistics Ph. D scientists who curate the literature Provide technical support to the team Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com
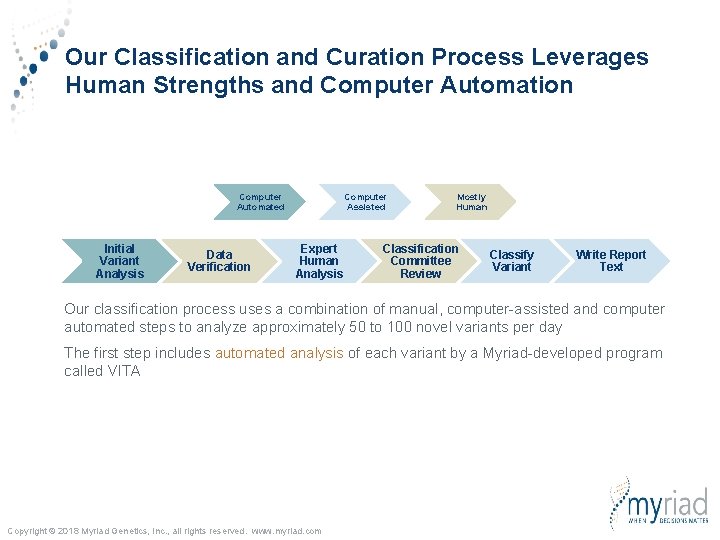
Our Classification and Curation Process Leverages Human Strengths and Computer Automation Computer Automated Initial Variant Analysis Data Verification Computer Assisted Expert Human Analysis Mostly Human Classification Committee Review Classify Variant Write Report Text Our classification process uses a combination of manual, computer-assisted and computer automated steps to analyze approximately 50 to 100 novel variants per day The first step includes automated analysis of each variant by a Myriad-developed program called VITA Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com
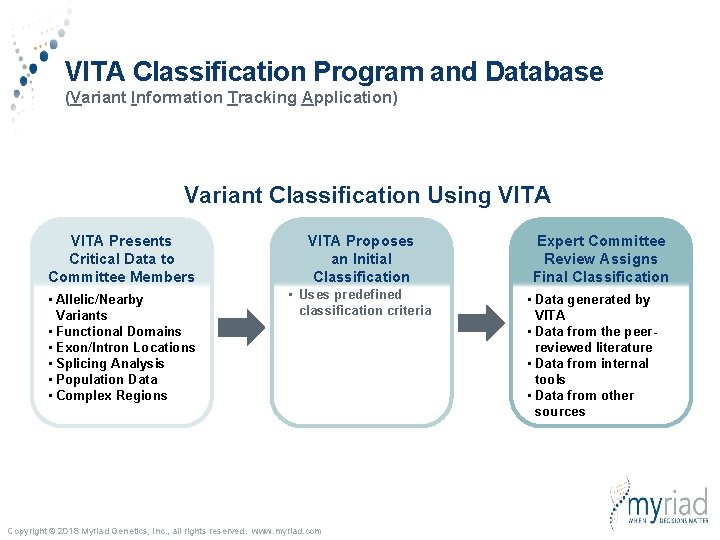
VITA Classification Program and Database (Variant Information Tracking Application) Variant Classification Using VITA Presents Critical Data to Committee Members VITA Proposes an Initial Classification • Allelic/Nearby Variants • Functional Domains • Exon/Intron Locations • Splicing Analysis • Population Data • Complex Regions • Uses predefined classification criteria Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com Expert Committee Review Assigns Final Classification • Data generated by VITA • Data from the peerreviewed literature • Data from internal tools • Data from other sources
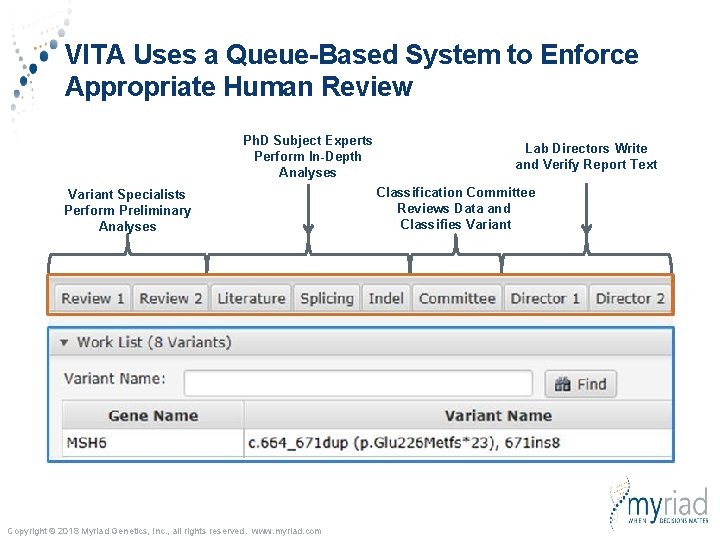
VITA Uses a Queue-Based System to Enforce Appropriate Human Review Ph. D Subject Experts Perform In-Depth Analyses Variant Specialists Perform Preliminary Analyses Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com Lab Directors Write and Verify Report Text Classification Committee Reviews Data and Classifies Variant
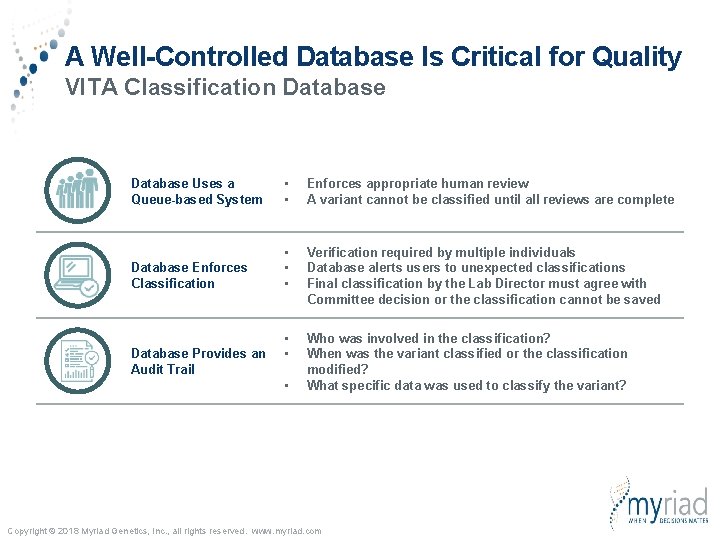
A Well-Controlled Database Is Critical for Quality VITA Classification Database Uses a Queue-based System • • Enforces appropriate human review A variant cannot be classified until all reviews are complete Database Enforces Classification • • • Verification required by multiple individuals Database alerts users to unexpected classifications Final classification by the Lab Director must agree with Committee decision or the classification cannot be saved • • Who was involved in the classification? When was the variant classified or the classification modified? What specific data was used to classify the variant? Database Provides an Audit Trail • Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com
Despite All of Our Classification Tools, VUS Are Unavoidable Question 1 Question 2 Question 3 Initial Variant Analysis What data quality and accuracy standards will we require? How will we maintain and document data integrity? How often will we update our database and variant classifications in order to meet patient needs? Data Verification Expert Human Analysis Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com Classification Committee Review VUS Write Report Text
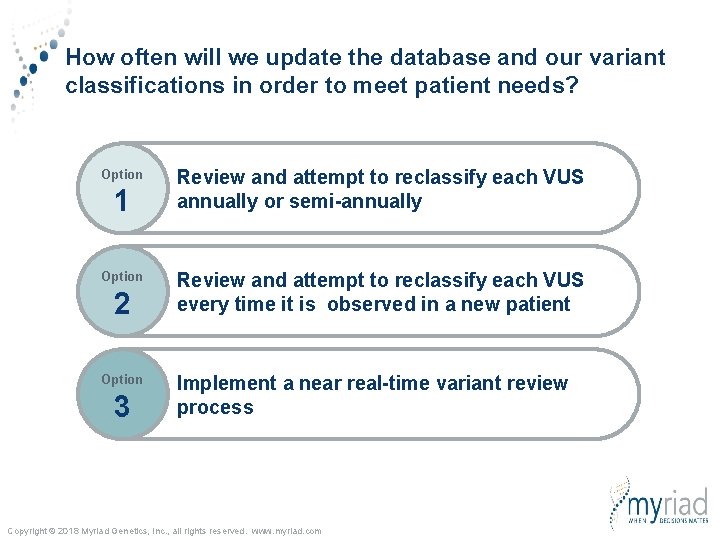
How often will we update the database and our variant classifications in order to meet patient needs? Option 1 Option 2 Option 3 Review and attempt to reclassify each VUS annually or semi-annually Review and attempt to reclassify each VUS every time it is observed in a new patient Implement a near real-time variant review process Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com
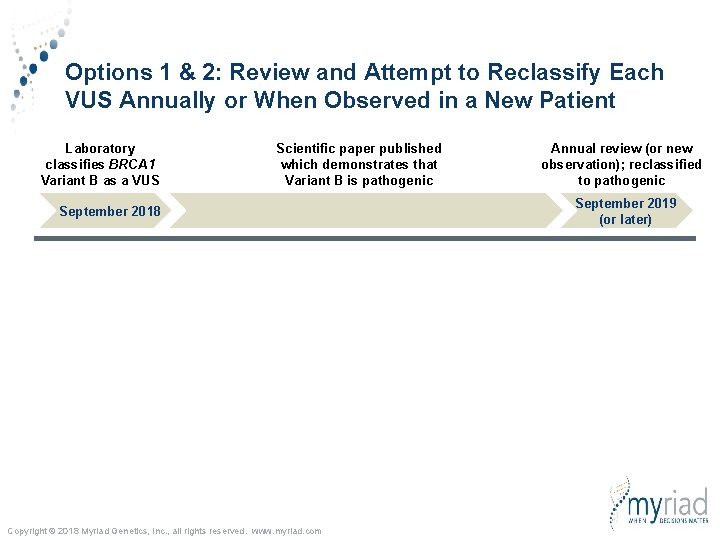
Options 1 & 2: Review and Attempt to Reclassify Each VUS Annually or When Observed in a New Patient Laboratory classifies BRCA 1 Variant B as a VUS Scientific paper published which demonstrates that Variant B is pathogenic September 2018 Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com Annual review (or new observation); reclassified to pathogenic September 2019 (or later)
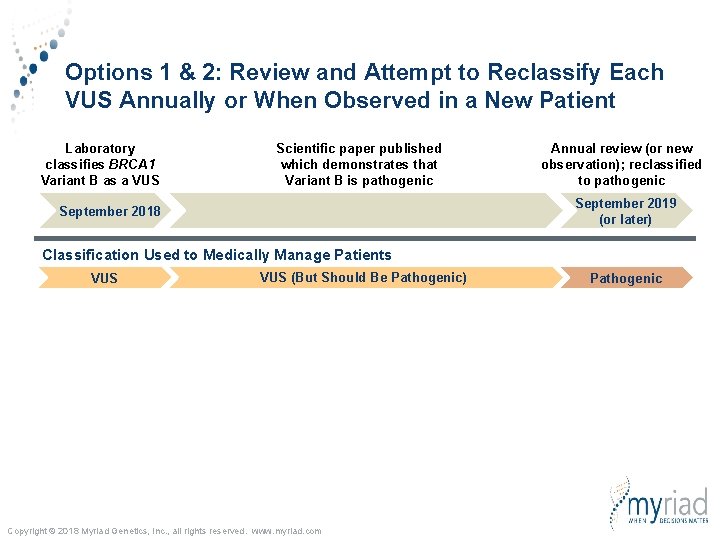
Options 1 & 2: Review and Attempt to Reclassify Each VUS Annually or When Observed in a New Patient Laboratory classifies BRCA 1 Variant B as a VUS Scientific paper published which demonstrates that Variant B is pathogenic Annual review (or new observation); reclassified to pathogenic September 2019 (or later) September 2018 Classification Used to Medically Manage Patients VUS (But Should Be Pathogenic) Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com Pathogenic
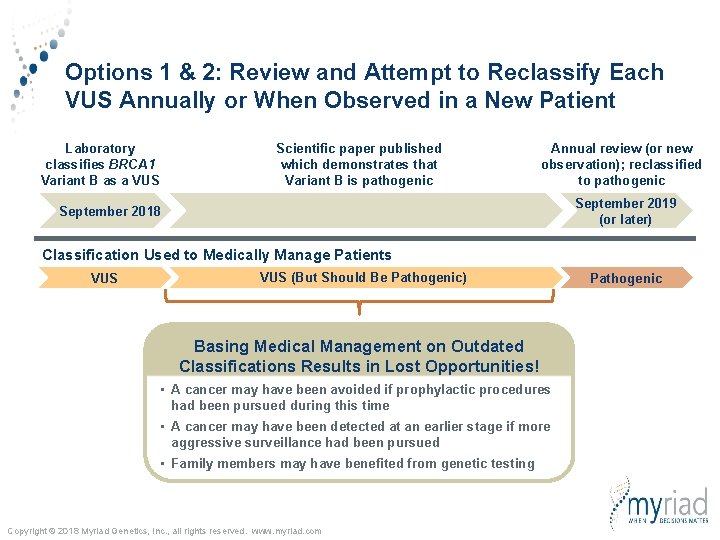
Options 1 & 2: Review and Attempt to Reclassify Each VUS Annually or When Observed in a New Patient Laboratory classifies BRCA 1 Variant B as a VUS Scientific paper published which demonstrates that Variant B is pathogenic Annual review (or new observation); reclassified to pathogenic September 2019 (or later) September 2018 Classification Used to Medically Manage Patients VUS (But Should Be Pathogenic) Basing Medical Management on Outdated Classifications Results in Lost Opportunities! • A cancer may have been avoided if prophylactic procedures had been pursued during this time • A cancer may have been detected at an earlier stage if more aggressive surveillance had been pursued • Family members may have benefited from genetic testing Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com Pathogenic
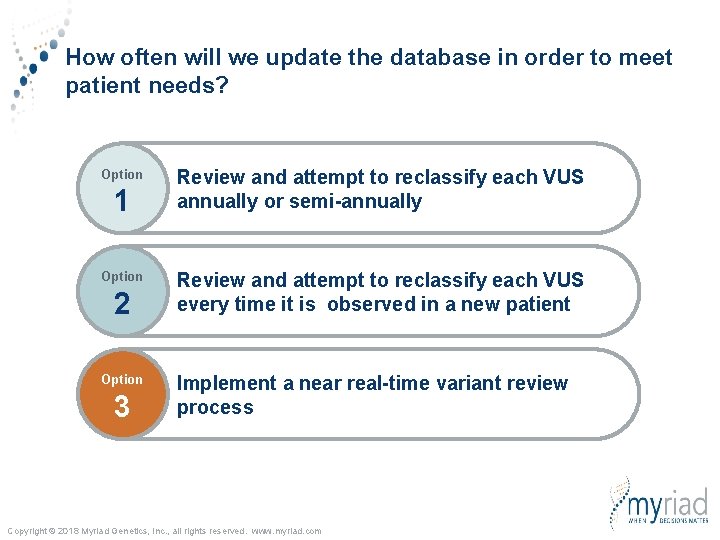
How often will we update the database in order to meet patient needs? Option 1 Option 2 Option 3 Review and attempt to reclassify each VUS annually or semi-annually Review and attempt to reclassify each VUS every time it is observed in a new patient Implement a near real-time variant review process Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com

Example 1: Literature is Reviewed in Real-Time Throughout the Lifetime of a Variant Before Test Launch Daily Literature Search First Observation (Classification) Daily Monitoring Before test launch, a complete literature search identifies previously reported variants, which are stored in our database with their associated papers. A daily literature search is performed by Ph. D-level scientists to keep our database current. Upon first observation of a variant at Myriad, targeted analysis verifies that critical papers were previously captured. Daily monitoring of the literature is performed in case new literature, which may allow us to reclassify a VUS, becomes available. Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com
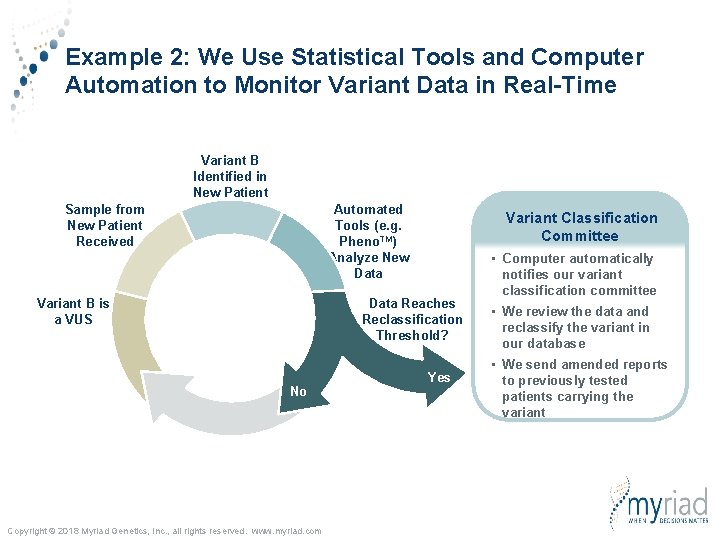
Example 2: We Use Statistical Tools and Computer Automation to Monitor Variant Data in Real-Time Variant B Identified in New Patient Sample from New Patient Received Automated Tools (e. g. Pheno. TM) Analyze New Data Variant B is a VUS Variant Classification Committee Data Reaches Reclassification Threshold? No Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com Yes • Computer automatically notifies our variant classification committee • We review the data and reclassify the variant in our database • We send amended reports to previously tested patients carrying the variant
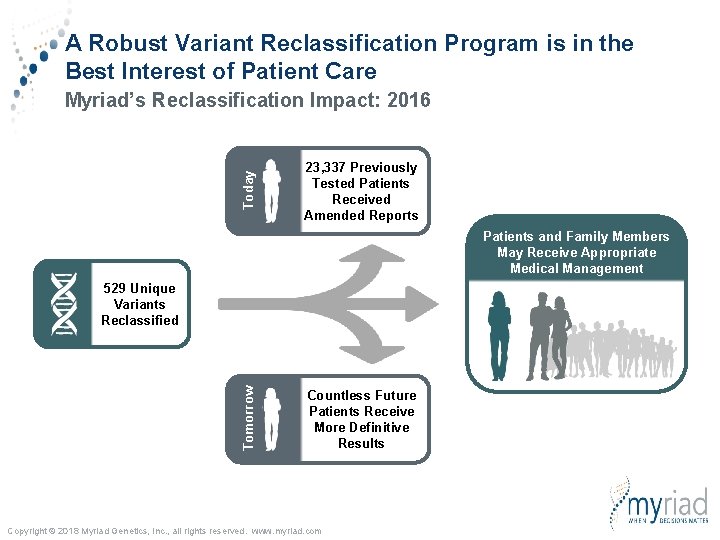
A Robust Variant Reclassification Program is in the Best Interest of Patient Care Today Myriad’s Reclassification Impact: 2016 23, 337 Previously Tested Patients Received Amended Reports Patients and Family Members May Receive Appropriate Medical Management Tomorrow 529 Unique Variants Reclassified Countless Future Patients Receive More Definitive Results Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com
Summary: Myriad’s Approach to a Clinical Database Our Data Must be of High Quality • • • Established high variant classification confidence thresholds Unbiased statistical tools used whenever possible Expert variant classification committee Database Integrity Must be Maintained • • Our database provides full traceability Who? When? and What specific data was used to classify or reclassify a variant? • • Development of innovative reclassification tools Near real-time monitoring of the scientific literature Automation of statistical analyses Notification of healthcare providers regarding variant reclassifications through amended patient reports Our Database Must Support Ongoing Variant Monitoring and Reclassification and the Issuance of Amended Reports Copyright © 2018 Myriad Genetics, Inc. , all rights reserved. www. myriad. com
- Slides: 31