Benjamin A Pierce GENETICS A Conceptual Approach SIXTH
Benjamin A. Pierce GENETICS A Conceptual Approach SIXTH EDITION CHAPTER 14 RNA Molecules and RNA Processing © 2017 W. H. Freeman and Company

Gene Organization • The concept of colinearity and noncolinearity • Number of nucleotides in a gene should be proportional to the number of amino acids in the encoded protein • DNA much longer than m. RNA; demonstrated through hybridization
Gene Organization • Introns • Exons
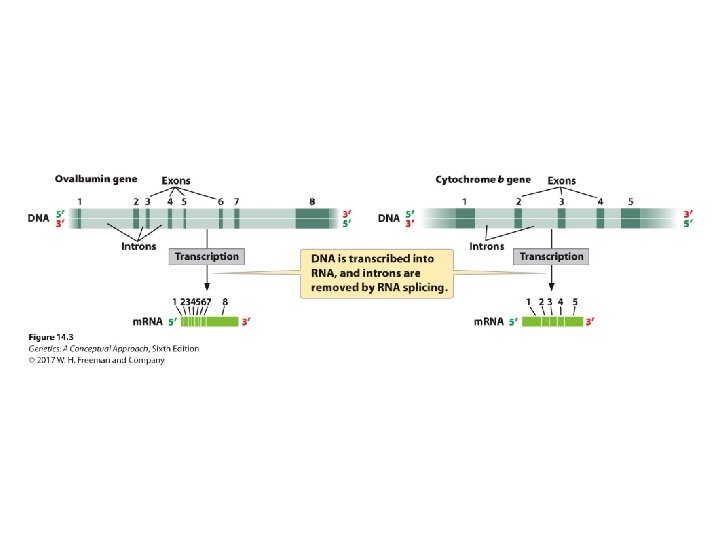
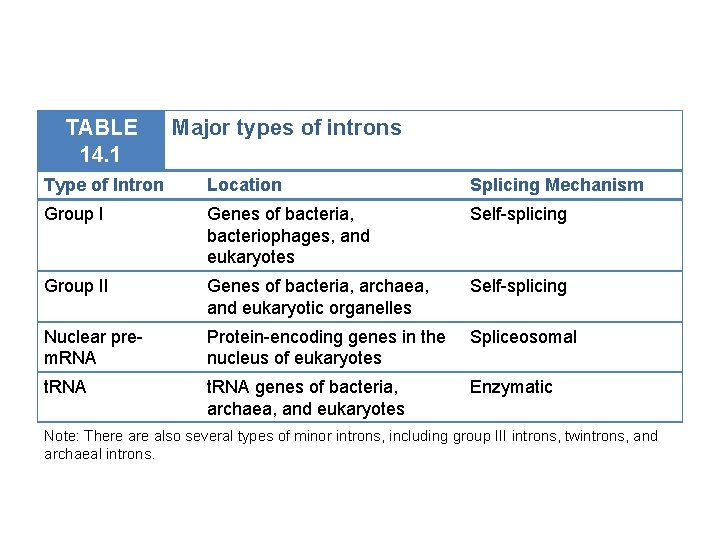
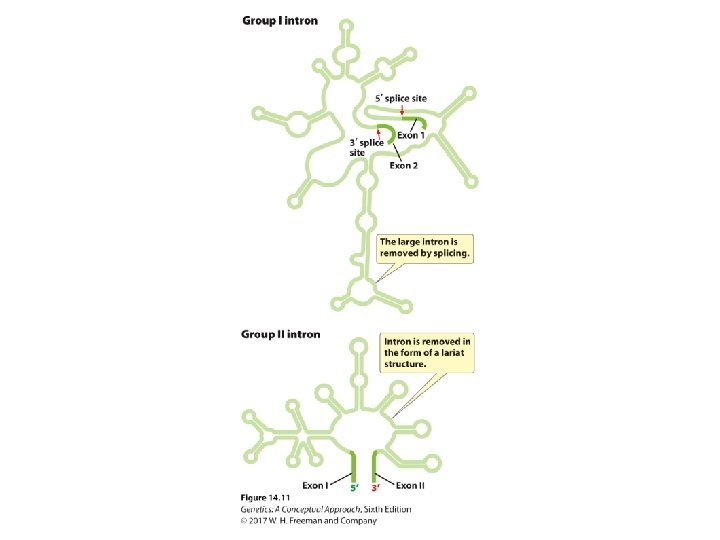
TABLE 14. 1 Major types of introns Type of Intron Location Splicing Mechanism Group I Genes of bacteria, bacteriophages, and eukaryotes Self-splicing Group II Genes of bacteria, archaea, and eukaryotic organelles Self-splicing Nuclear prem. RNA Protein-encoding genes in the nucleus of eukaryotes Spliceosomal t. RNA genes of bacteria, archaea, and eukaryotes Enzymatic Note: There also several types of minor introns, including group III introns, twintrons, and archaeal introns.
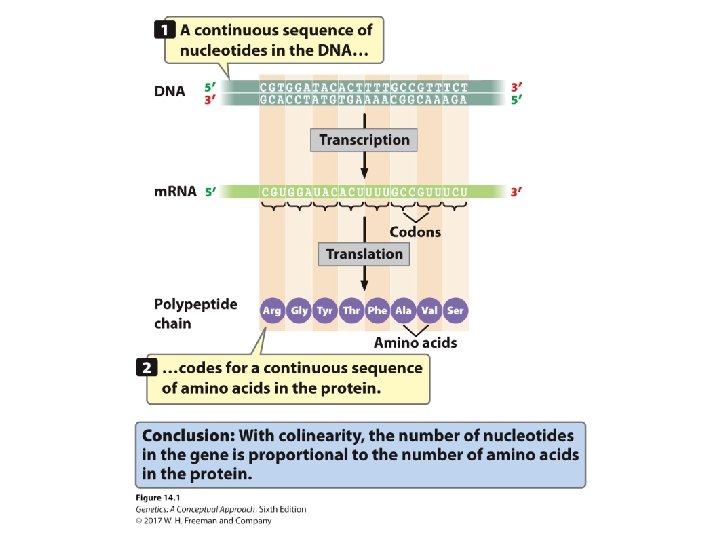
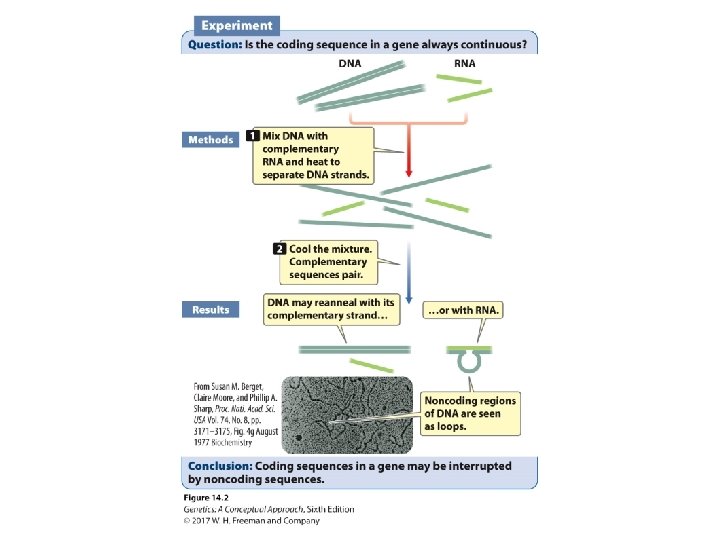
The Concept of the Gene The gene includes • DNA sequences that code for all exons and introns • Those sequences at the beginning and end of the RNA that are not translated into a protein, including the entire transcription unit – The promoter – The RNA coding sequence – The terminator
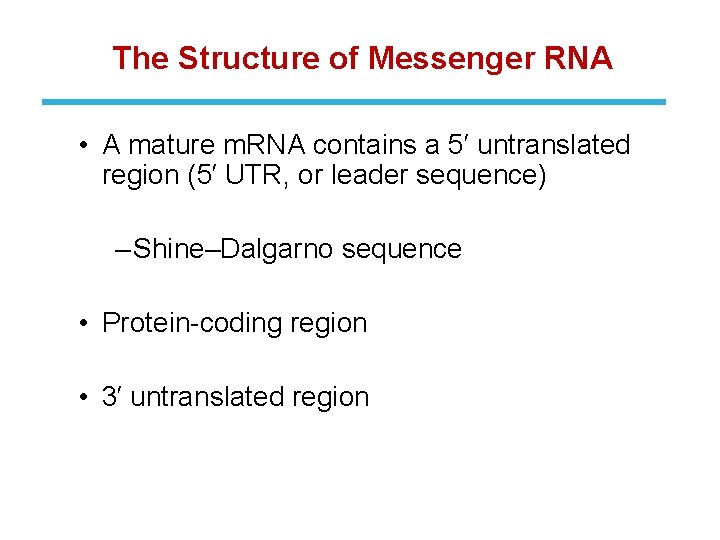
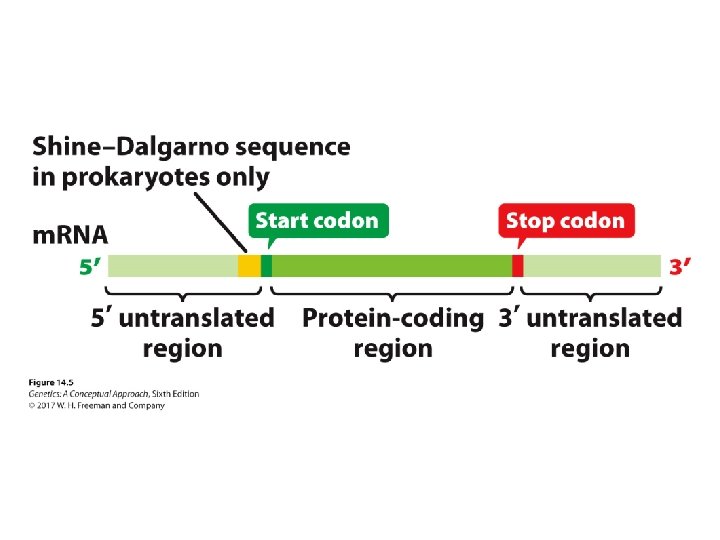
The Structure of Messenger RNA • A mature m. RNA contains a 5′ untranslated region (5′ UTR, or leader sequence) ‒ Shine–Dalgarno sequence • Protein-coding region • 3′ untranslated region
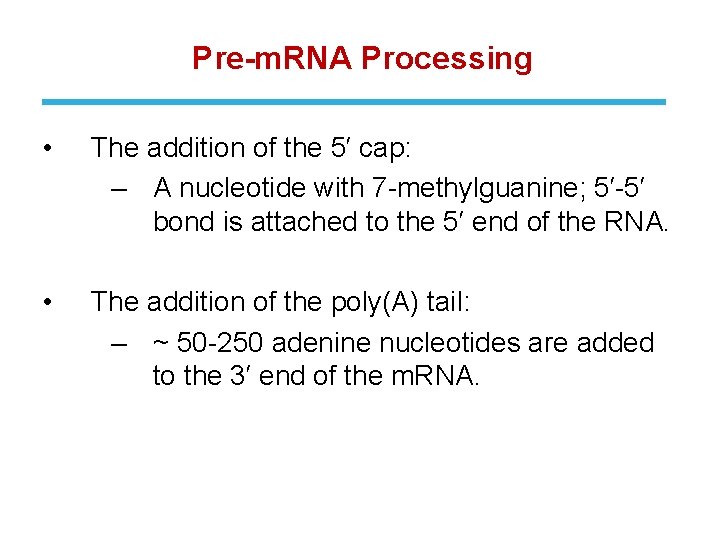
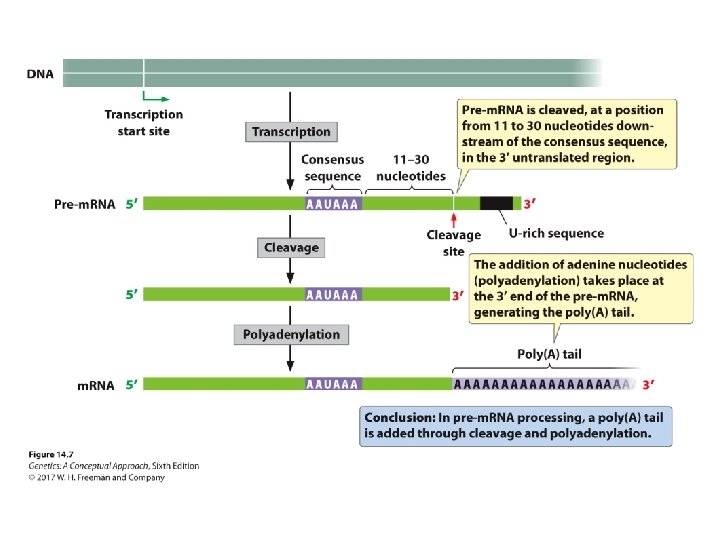
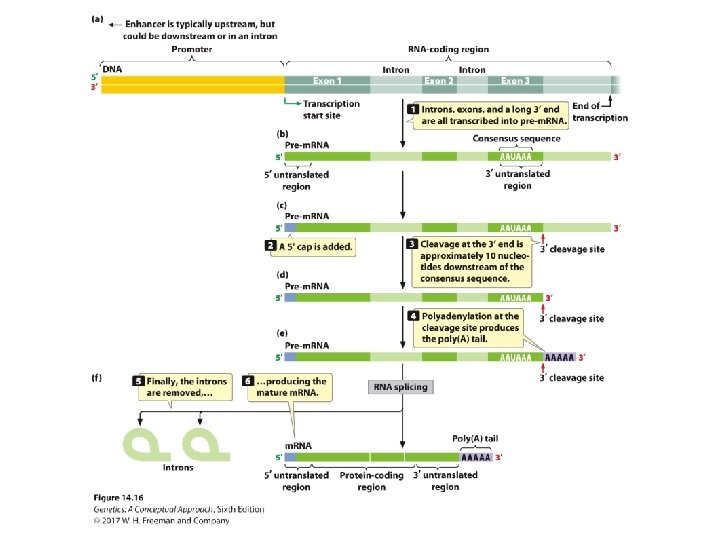
Pre-m. RNA Processing • The addition of the 5′ cap: ‒ A nucleotide with 7 -methylguanine; 5′-5′ bond is attached to the 5′ end of the RNA. • The addition of the poly(A) tail: ‒ ~ 50 -250 adenine nucleotides are added to the 3′ end of the m. RNA.
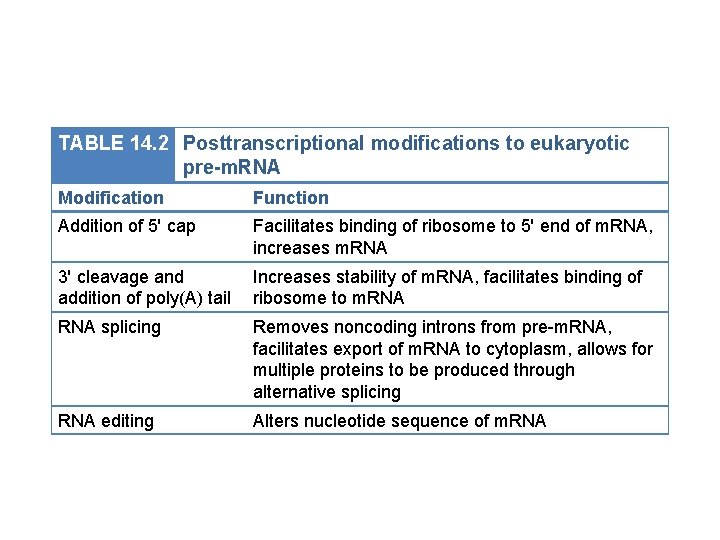
TABLE 14. 2 Posttranscriptional modifications to eukaryotic pre-m. RNA Modification Function Addition of 5' cap Facilitates binding of ribosome to 5' end of m. RNA, increases m. RNA 3' cleavage and addition of poly(A) tail Increases stability of m. RNA, facilitates binding of ribosome to m. RNA splicing Removes noncoding introns from pre-m. RNA, facilitates export of m. RNA to cytoplasm, allows for multiple proteins to be produced through alternative splicing RNA editing Alters nucleotide sequence of m. RNA
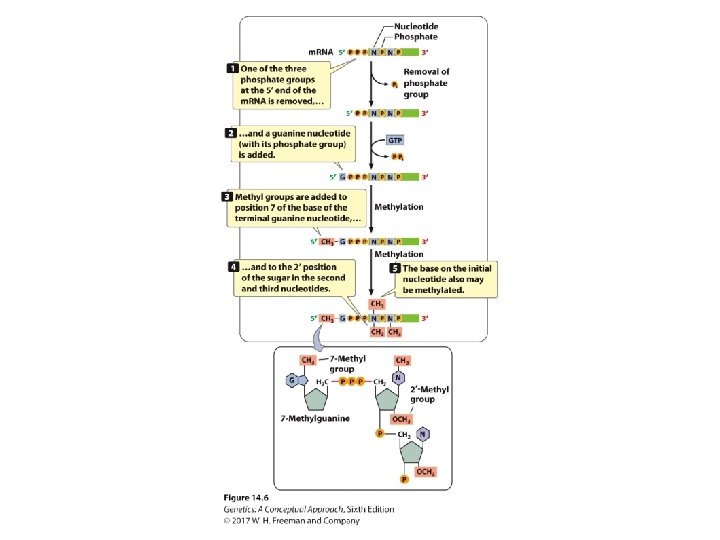
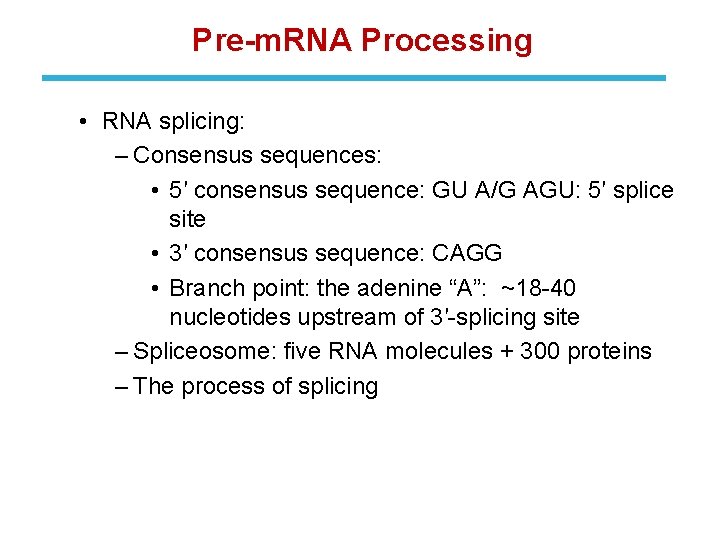
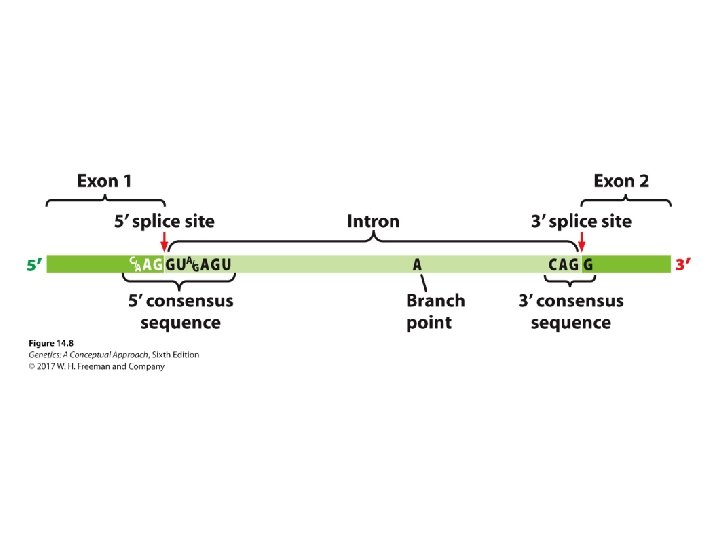
Pre-m. RNA Processing • RNA splicing: ‒ Consensus sequences: • 5′ consensus sequence: GU A/G AGU: 5′ splice site • 3′ consensus sequence: CAGG • Branch point: the adenine “A”: ~18 -40 nucleotides upstream of 3′-splicing site ‒ Spliceosome: five RNA molecules + 300 proteins ‒ The process of splicing
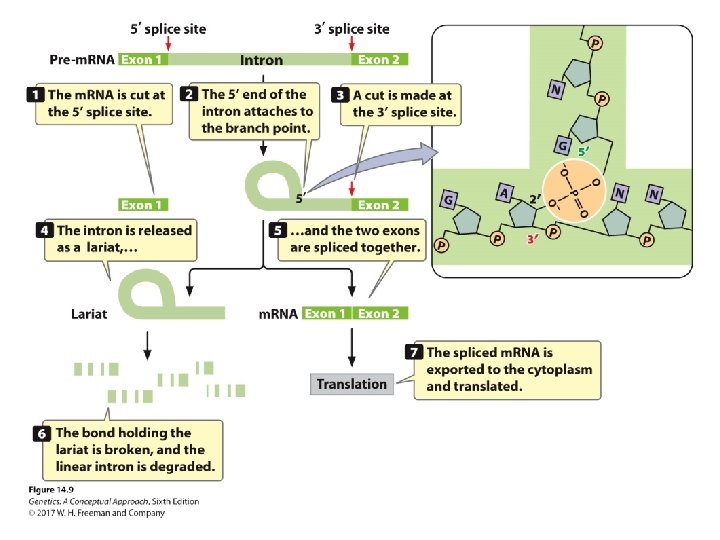
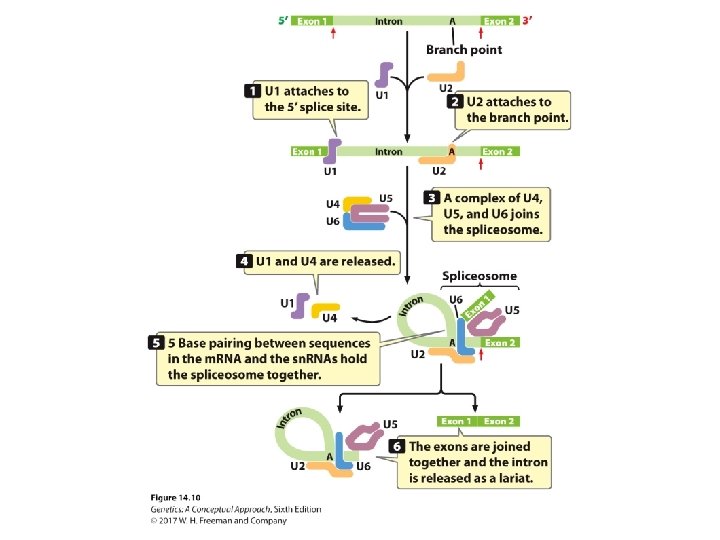

Concept Check 1 If a splice site were mutated so that splicing did not take place, what would the effect be on the protein encoded by the m. RNA? a. It would be shorter than normal. b. It would be longer than normal. c. It would be the same length but would have different amino acids.

Concept Check 1 If a splice site were mutated so that splicing did not take place, what would the effect be on the protein encoded by the m. RNA? a. It would be shorter than normal. b. It would be longer than normal. c. It would be the same length but would have different amino acids.
Pre-m. RNA Processing • Nuclear organization • Intron removal, m. RNA processing, and transcription take place at the same site in the nucleus. • Minor splicing • Self-splicing introns in some r. RNA genes in protists and in mitochondria genes in fungi • Alternative processing pathways for processing prem. RNA
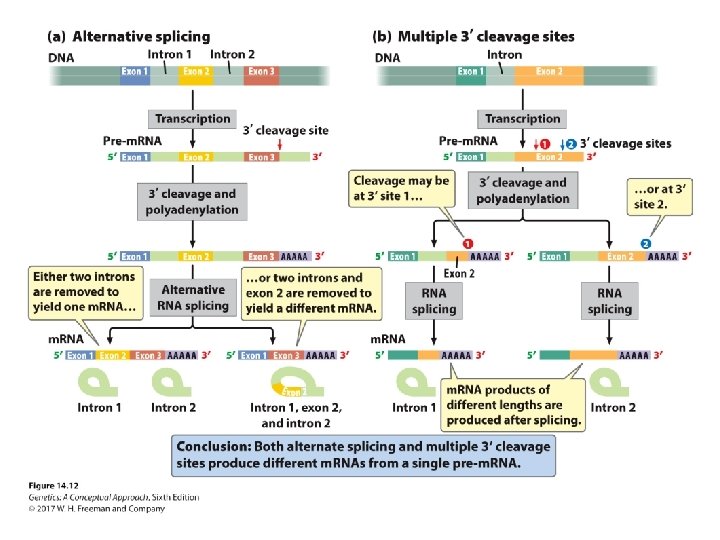
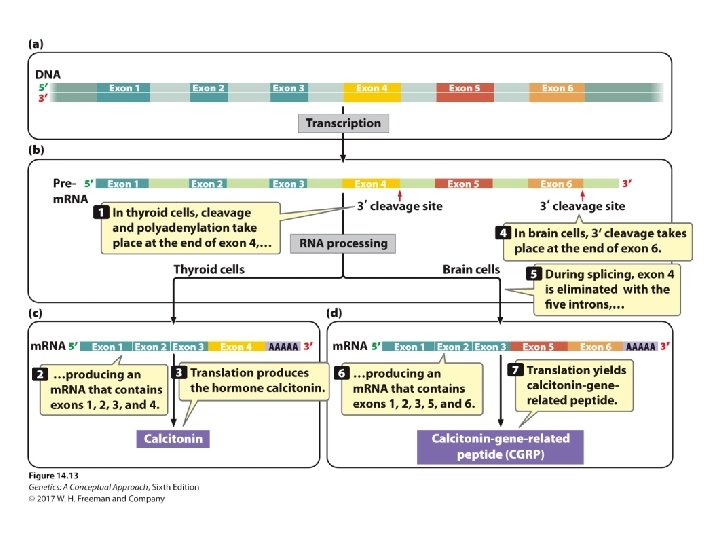
Concept Check 2 Alternative 3′ cleavage sites result in _____. a. b. c. d. multiple genes of different lengths multiple genes of pre-m. RNA of different lengths multiple m. RNAs of different lengths All of the above
Concept Check 2 Alternative 3′ cleavage sites result in _____. a. b. c. d. multiple genes of different lengths multiple genes of pre-m. RNA of different lengths multiple m. RNAs of different lengths All of the above

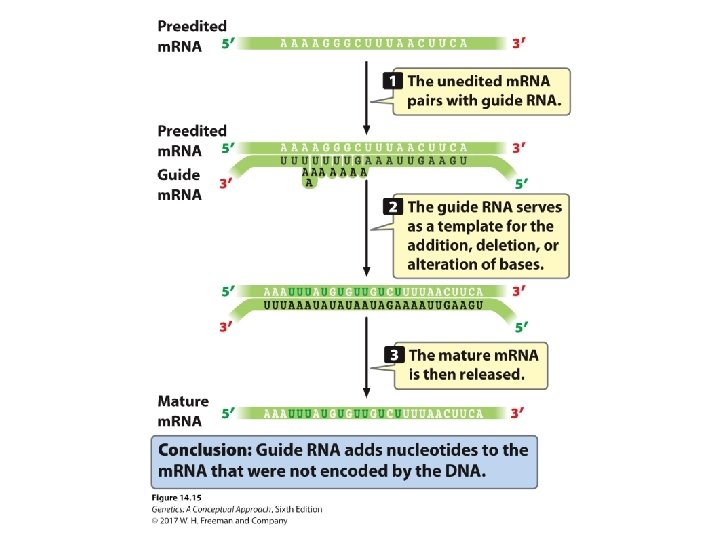
RNA Editing RNA editing: coding sequence altered after transcription. Guide RNAs play a role.

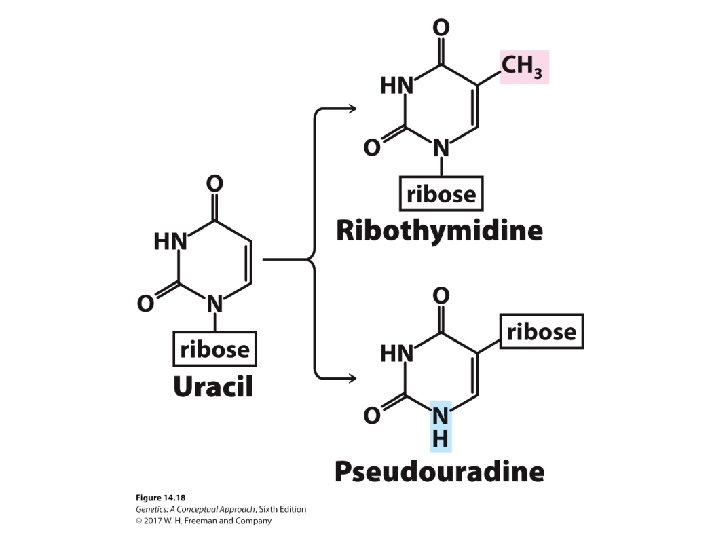
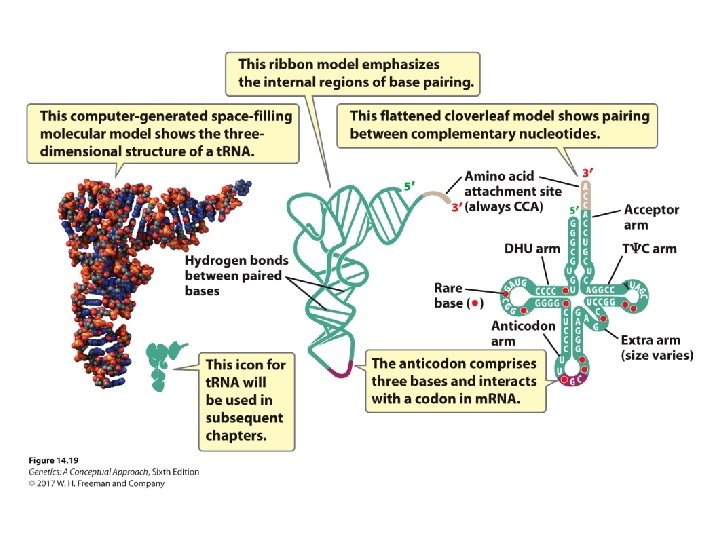
The Structure of Transfer RNA • Rare modified RNA nucleotide bases ‒ Ribothymine ‒ Pseudouridine • Common secondary structure—the cloverleaf structure • Anticodon
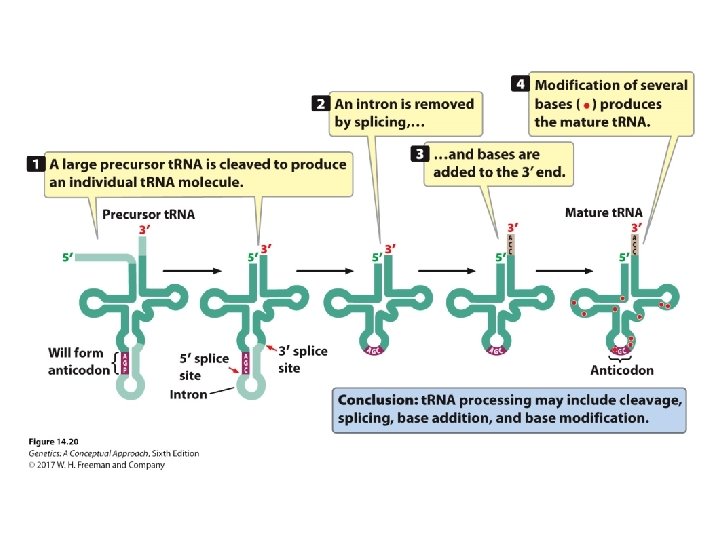
Concept Check 3 How are rare bases incorporated into t. RNAs? a. b. c. d. Encoded by guide RNAs By chemical changes in one of the standard bases Encoded by rare bases in DNA Encoded by sequences in introns
Concept Check 3 How are rare bases incorporated into t. RNAs? a. b. c. d. Encoded by guide RNAs By chemical changes in one of the standard bases Encoded by rare bases in DNA Encoded by sequences in introns

The Structure of the Ribosome • Large ribosome subunit • Small ribosome subunit
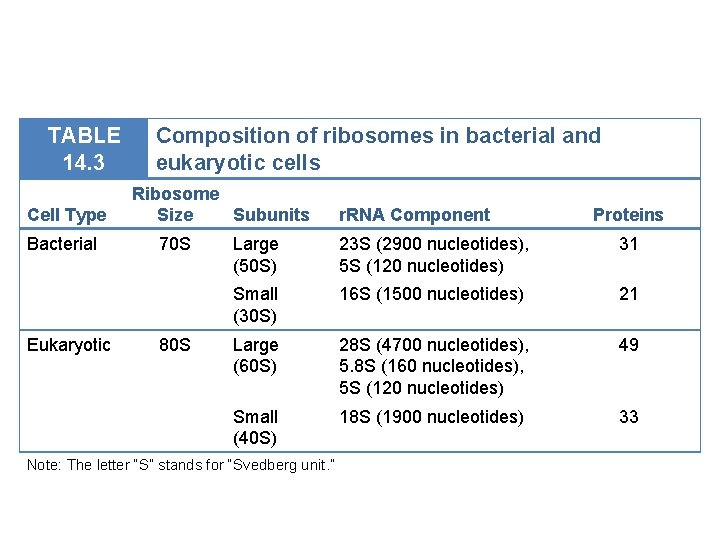
TABLE 14. 3 Cell Type Bacterial Eukaryotic Composition of ribosomes in bacterial and eukaryotic cells Ribosome Size Subunits 70 S 80 S r. RNA Component Proteins Large (50 S) 23 S (2900 nucleotides), 5 S (120 nucleotides) 31 Small (30 S) 16 S (1500 nucleotides) 21 Large (60 S) 28 S (4700 nucleotides), 5. 8 S (160 nucleotides), 5 S (120 nucleotides) 49 Small (40 S) 18 S (1900 nucleotides) 33 Note: The letter “S” stands for “Svedberg unit. ”
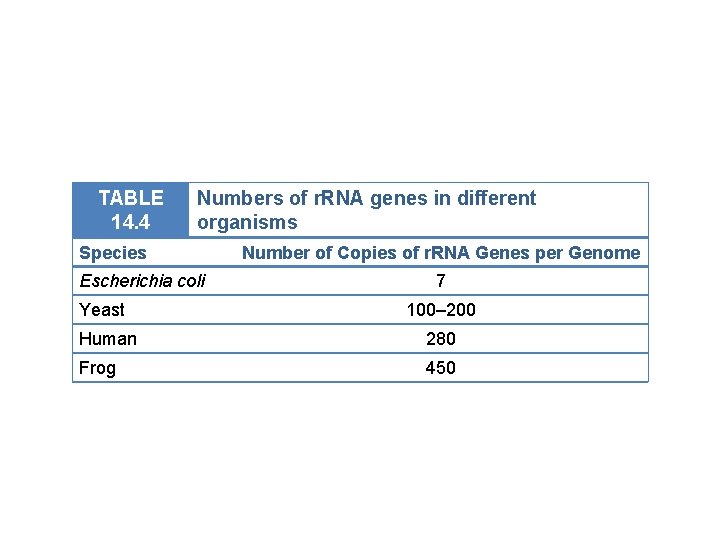
TABLE 14. 4 Numbers of r. RNA genes in different organisms Species Escherichia coli Yeast Number of Copies of r. RNA Genes per Genome 7 100– 200 Human 280 Frog 450
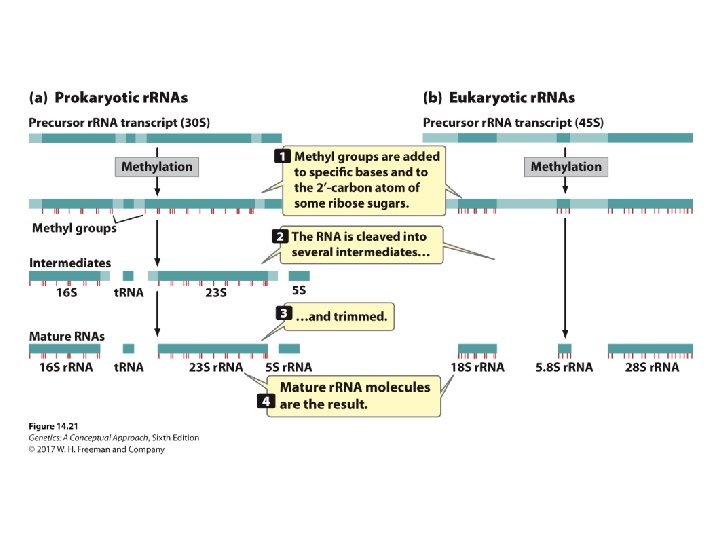
Concept Check 4 What types of changes take place in r. RNA processing? a. b. c. d. Methylation of bases Cleavage of bases Nucleotides trimmed from the ends of r. RNAs All of the above
Concept Check 4 What types of changes take place in r. RNA processing? a. b. c. d. Methylation of bases Cleavage of bases Nucleotides trimmed from the ends of r. RNAs All of the above
14. 5 Small RNA Molecules Participate in a Variety of Functions • RNA interference: limits the invasion of foreign genes and censors the expression of their own genes • Types of small RNAs • Processing and function of micro. RNAs
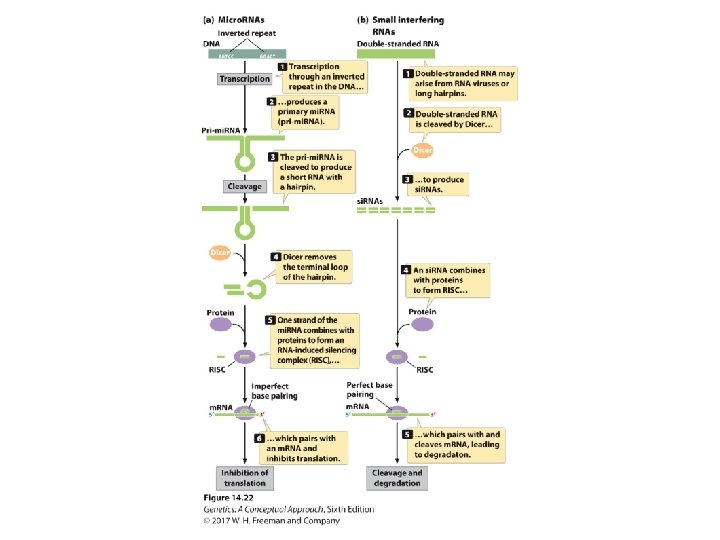
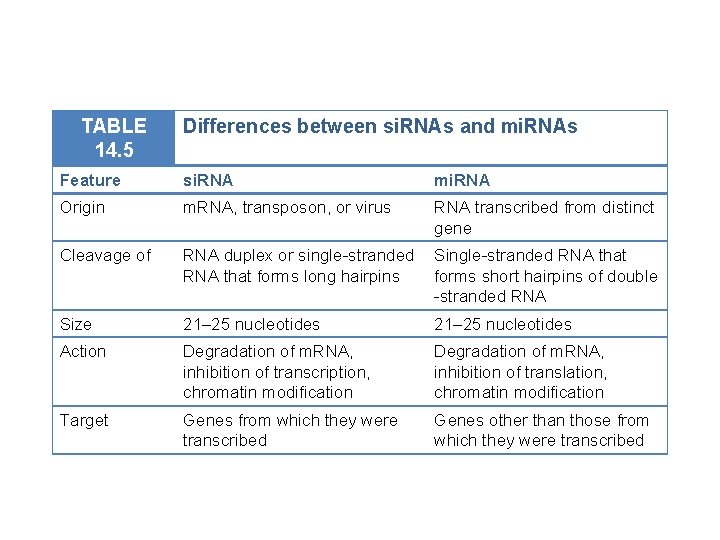
TABLE 14. 5 Differences between si. RNAs and mi. RNAs Feature si. RNA mi. RNA Origin m. RNA, transposon, or virus RNA transcribed from distinct gene Cleavage of RNA duplex or single-stranded RNA that forms long hairpins Single-stranded RNA that forms short hairpins of double -stranded RNA Size 21– 25 nucleotides Action Degradation of m. RNA, inhibition of transcription, chromatin modification Degradation of m. RNA, inhibition of translation, chromatin modification Target Genes from which they were transcribed Genes other than those from which they were transcribed
- Slides: 48