Benjamin A Pierce GENETICS A Conceptual Approach FIFTH

Benjamin A. Pierce GENETICS A Conceptual Approach FIFTH EDITION CHAPTER 15 The Genetic Code and Translation © 2014 W. H. Freeman and Company

15. 1 Many Genes Encode Proteins The One Gene, One Enzyme Hypothesis • Genes function by encoding enzymes, and each gene encodes a separate enzyme. • More specific: one gene, one polypeptide hypothesis.
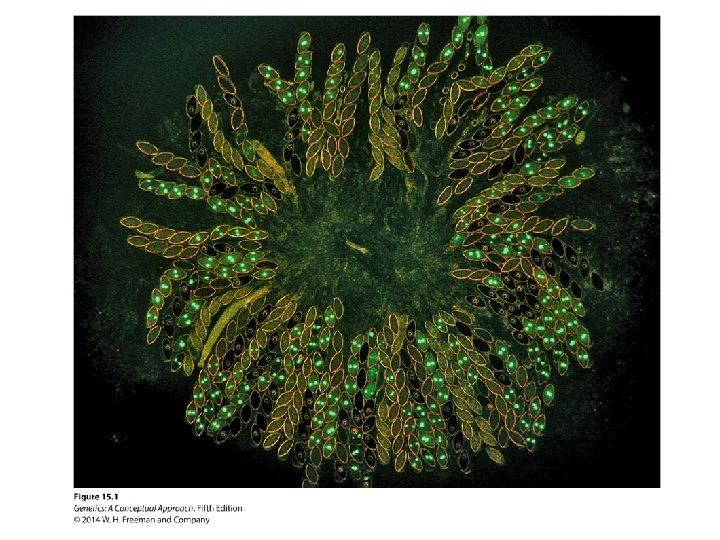

Concept Check 1 Auxotrophic mutation 103 grows on minimal medium supplemented with A, B, or C. Mutation 106 grows on medium supplemented with A and C, but not B; and mutation 102 grows only on medium supplemented with C. What is the order of A, B, C in a biochemical pathway?
Concept Check 1 Auxotrophic mutation 103 grows on minimal medium supplemented with A, B, or C. Mutation 106 grows on medium supplemented with A and C, but not B; and mutation 102 grows only on medium supplemented with C. What is the order of A, B, C in a biochemical pathway? B A C
15. 1 Many Genes Encode Proteins The Structure and Function of Proteins • Proteins are polymers consisting of amino acids linked by peptide bonds. • Amino acid sequence is its primary structure. • This structure folds to create secondary and tertiary structures. • Two or more polypeptide chains associate to form quaternary structure.

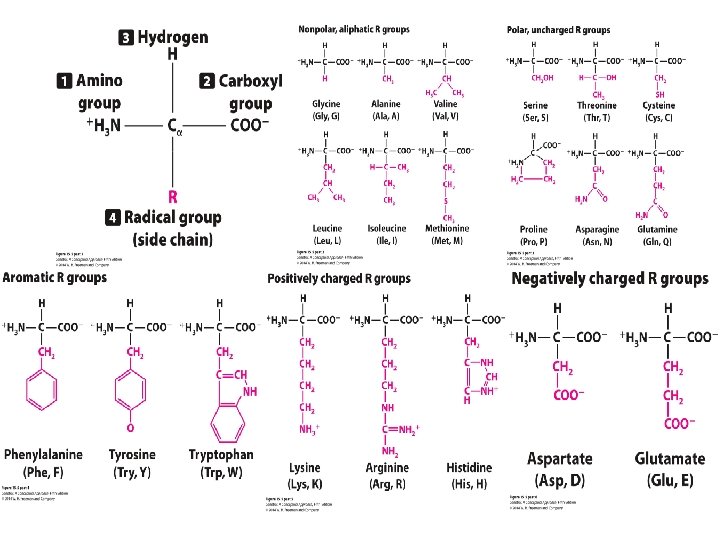
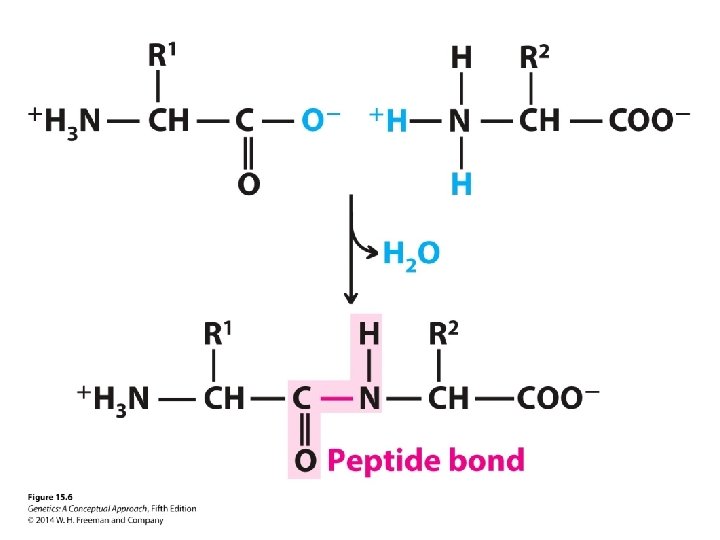
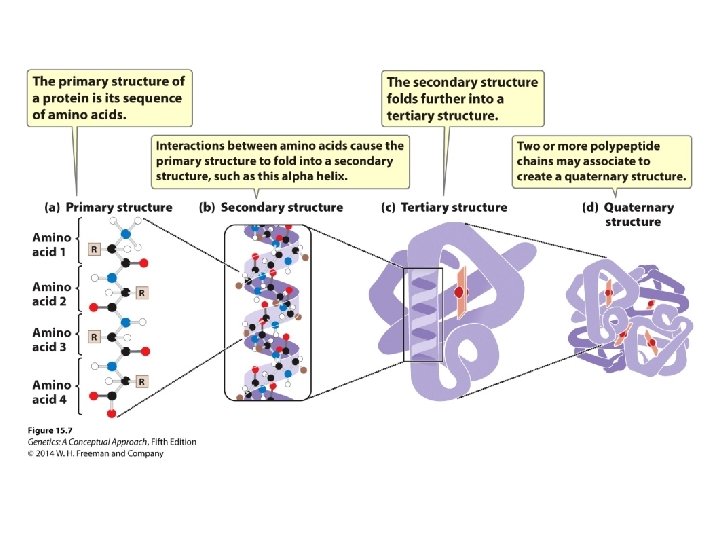

Concept Check 2 What primarily determines the secondary and tertiary structures of a protein?

Concept Check 2 What primarily determines the secondary and tertiary structures of a protein? The primary structure
15. 2 The Genetic Code Determines How the Nucleotide Sequence Specifies the Amino Acid Sequence of a Protein • Breaking the Genetic Code • Degeneracy of the Code • The Reading Frame and Initiation Codons • Termination Codons • The Universality of the Code
Breaking the Genetic Code • Homopolymers • Random copolymers • Ribosome-bound t. RNAs
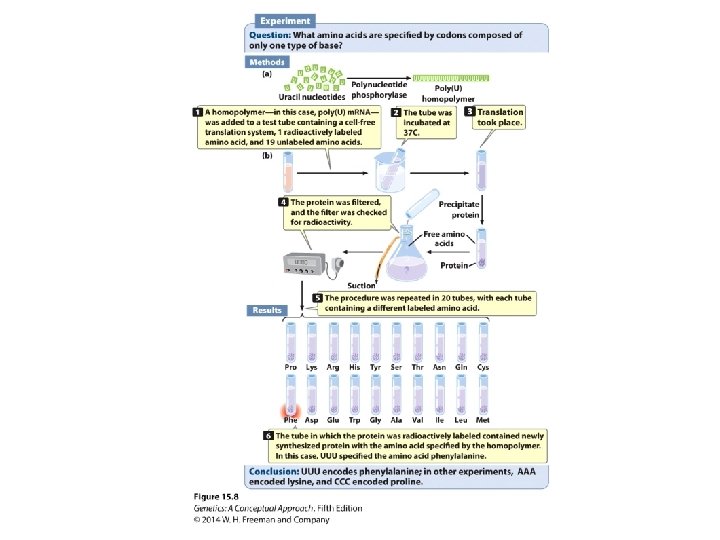
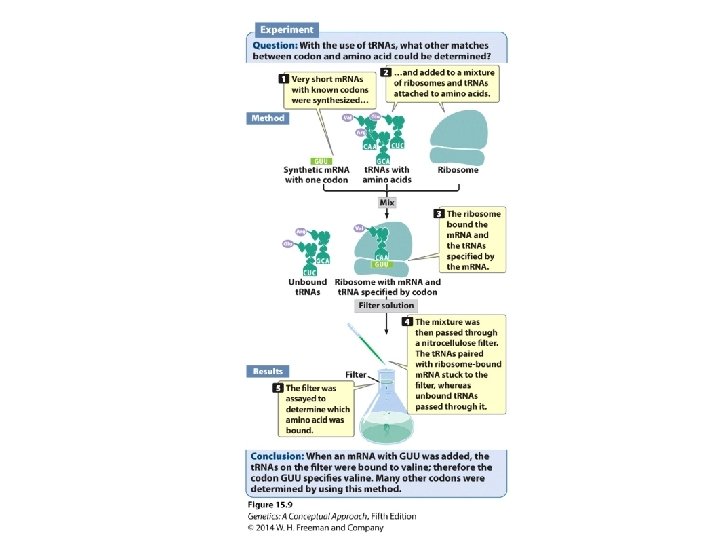

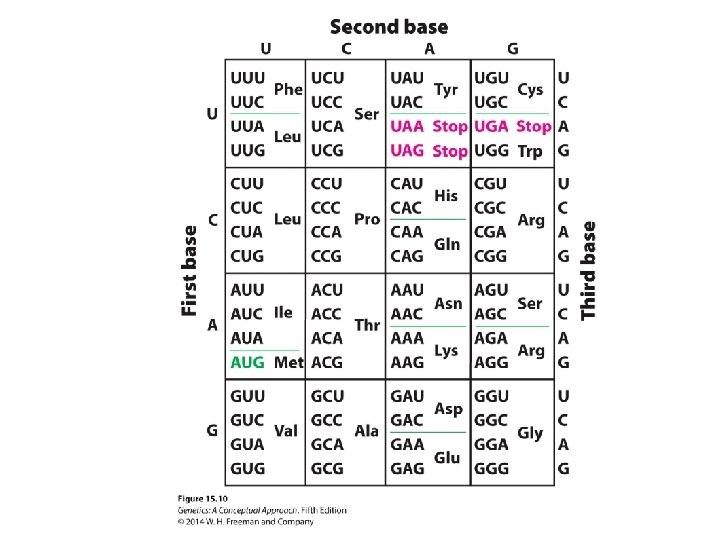
Breaking the Genetic Code • Codon: a triplet RNA code • 64 possible codons: ‒ 3 stop codons ‒ 61 sense codons
The Degeneracy of the Code • Degenerate code: amino acid may be specified by more than one codon. • Synonymous codons: codons that specify the same amino acid • Isoaccepting t. RNAs: different t. RNAs that accept the same amino acid but have different anticodons
The Degeneracy of the Code • Codons – Sense codons: encoding amino acid – Initiation codon: AUG – Termination codon: UAA, UAG, UGA • Wobble hypothesis
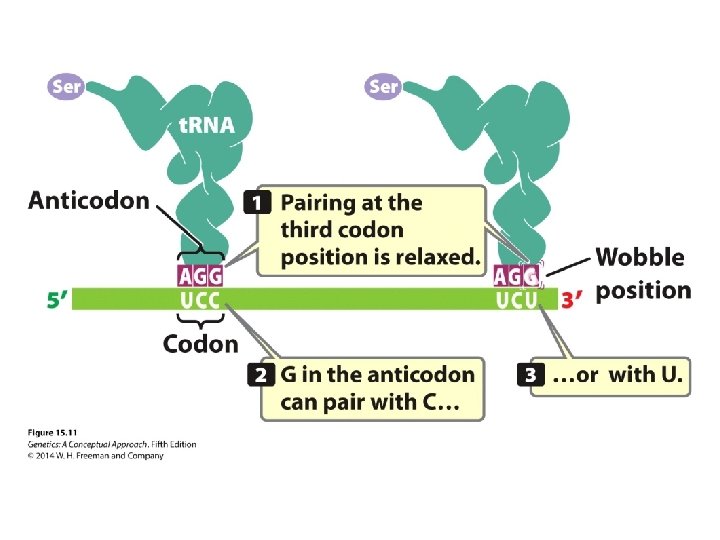
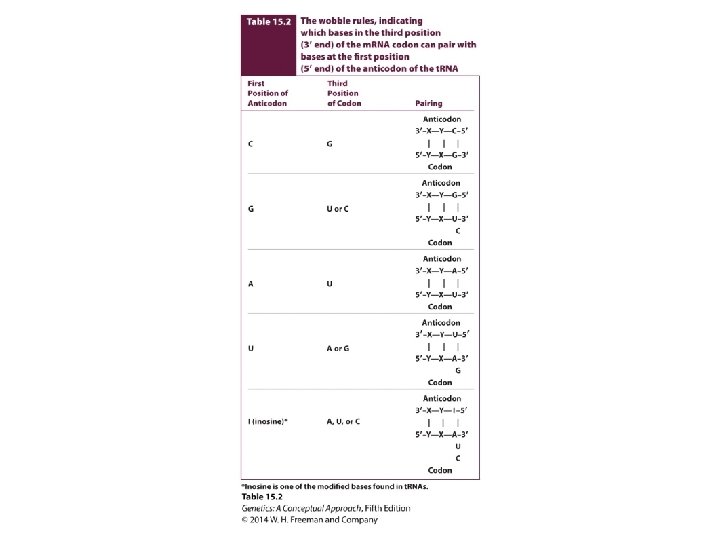
Concept Check 3 Through wobble, a single than one. can pair with more a. codon; anticodon b. group of three nucleotides in DNA; codon in m. RNA c. t. RNA; amino acid d. anticodon; codon
Concept Check 3 Through wobble, a single than one. can pair with more a. codon; anticodon b. group of three nucleotides in DNA; codon in m. RNA c. t. RNA; amino acid d. anticodon; codon
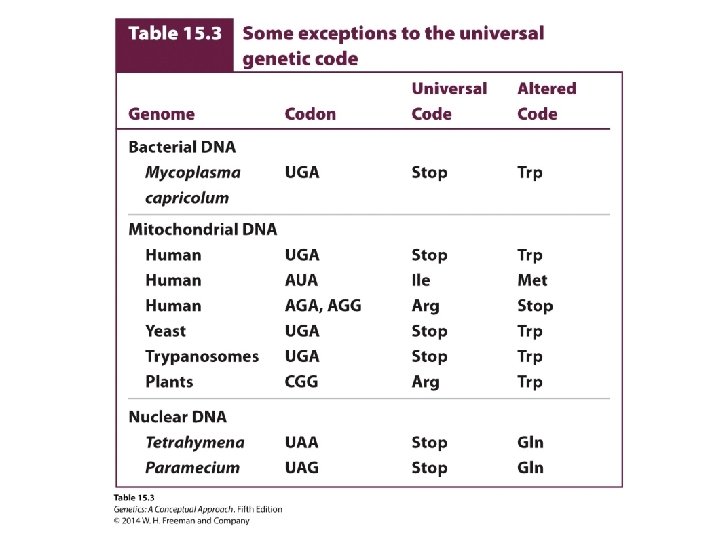
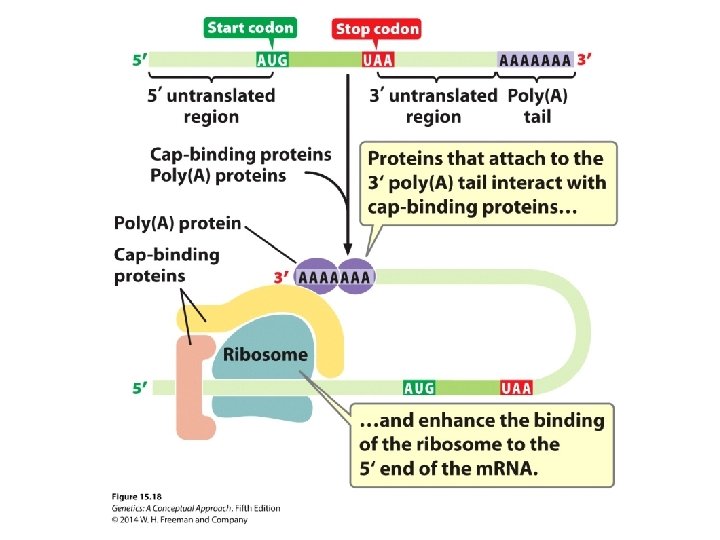
The Reading Frame and Initiation Codons • Reading frame: three ways in which the sequence can be read in groups of three. Each different way of reading encodes a different amino acid sequence. • Nonoverlapping: A single nucleotide may not be included in more than one codon. • The universality of the code: near universal, with some exceptions
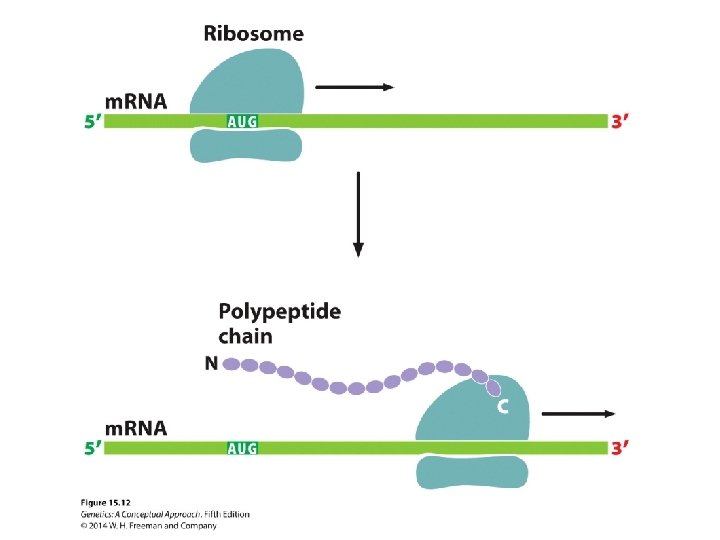
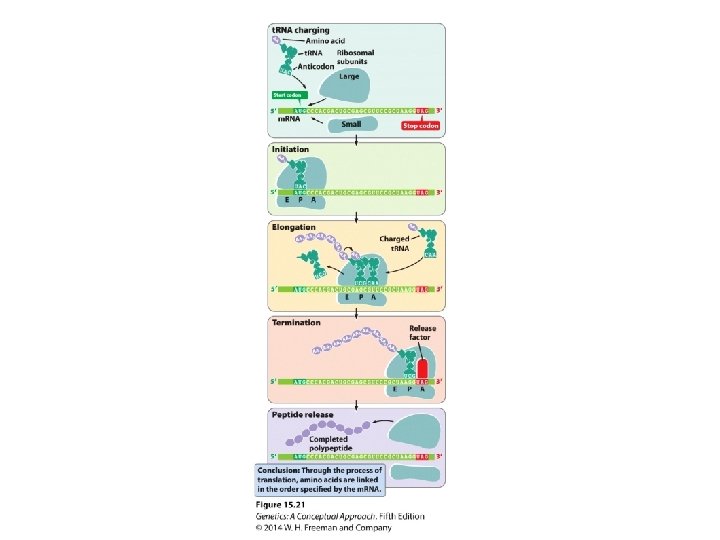
15. 3 Amino Acids Are Assembled into a Protein Through Translation • The binding of amino acids to transfer RNAs • The initiation of translation • Elongation • Termination
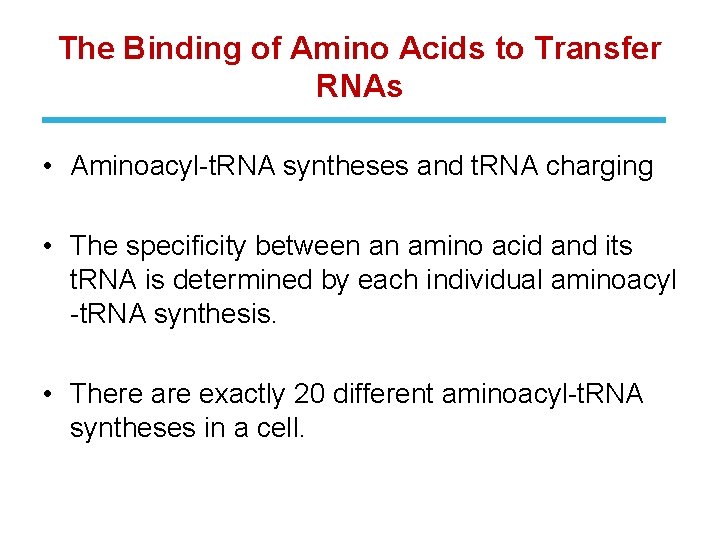
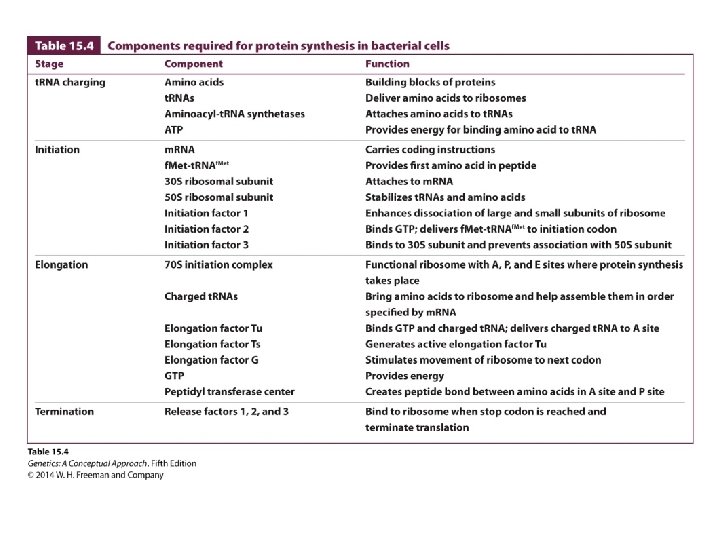
The Binding of Amino Acids to Transfer RNAs • Aminoacyl-t. RNA syntheses and t. RNA charging • The specificity between an amino acid and its t. RNA is determined by each individual aminoacyl -t. RNA synthesis. • There are exactly 20 different aminoacyl-t. RNA syntheses in a cell.
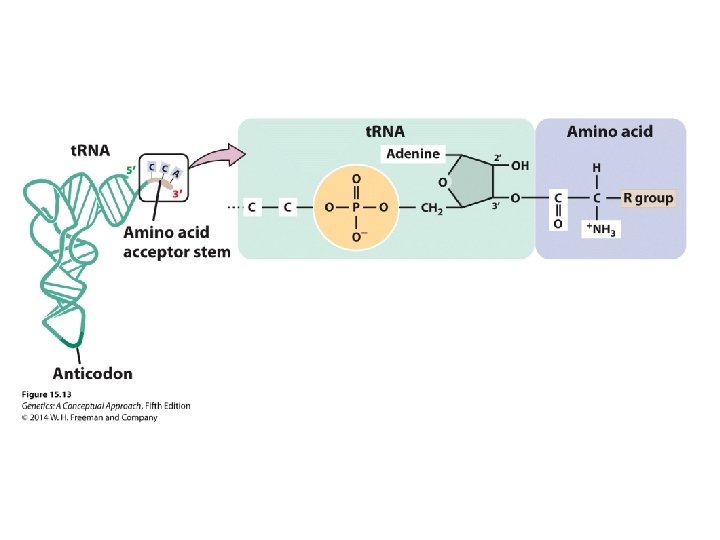
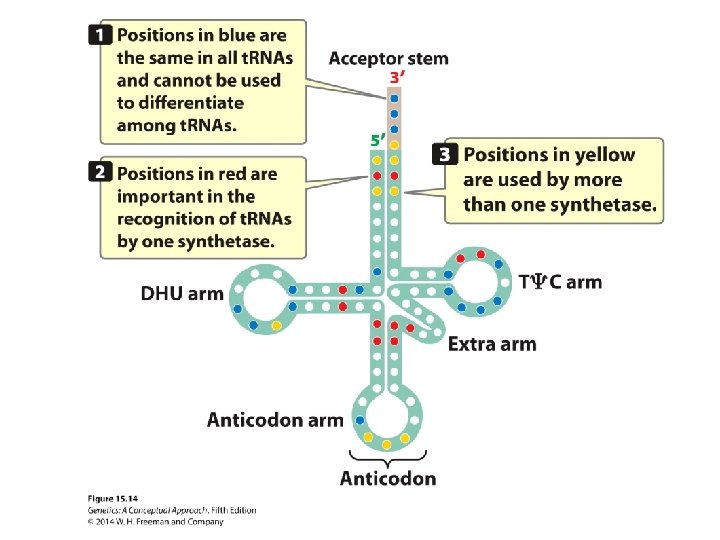
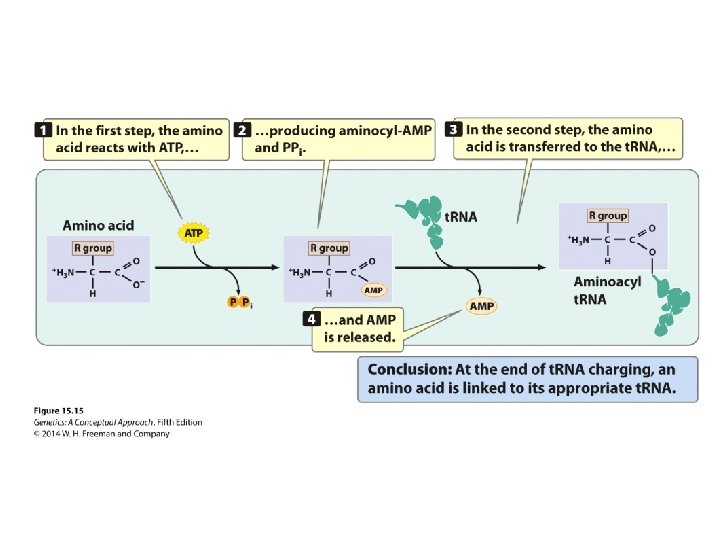
Concept Check 4 Amino acids bind to which part of the t. RNA? a. b. c. d. anticodon DHU arm 3′ end 5′ end
Concept Check 4 Amino acids bind to which part of the t. RNA? a. b. c. d. anticodon DHU arm 3′ end 5′ end
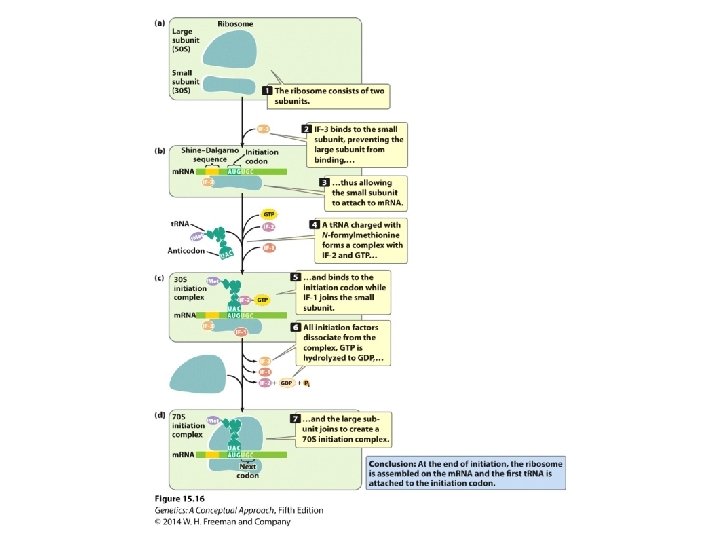
The Initiation of Translation • Initiation factors IF-3, initiator t. RNA with Nformylmethionine attached to form fmet-t. RNA • Energy molecule: GTP
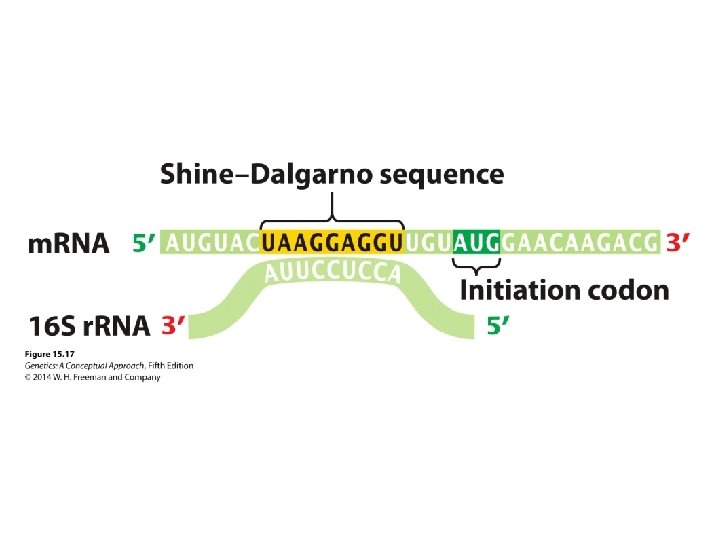
The Initiation of Translation • The Shine–Dalgarno consensus sequence in bacterial cells is recognized by the small unit of ribosome. • The Kozak sequence in eukaryotic cells facilitates the identification of the start codon.
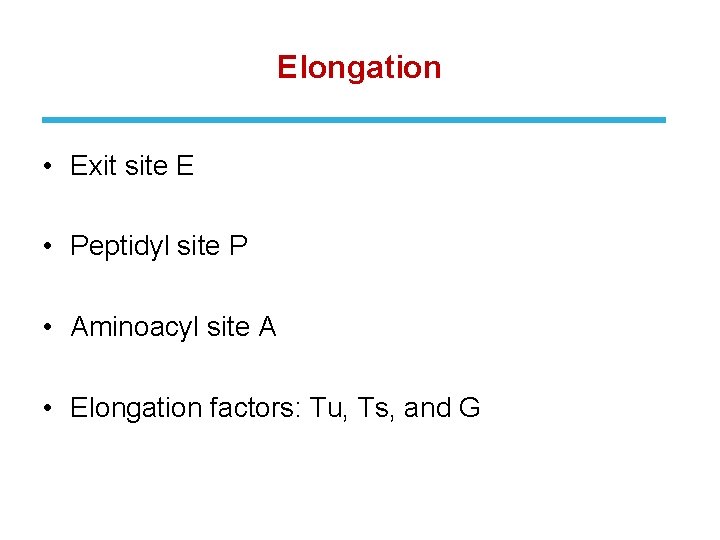
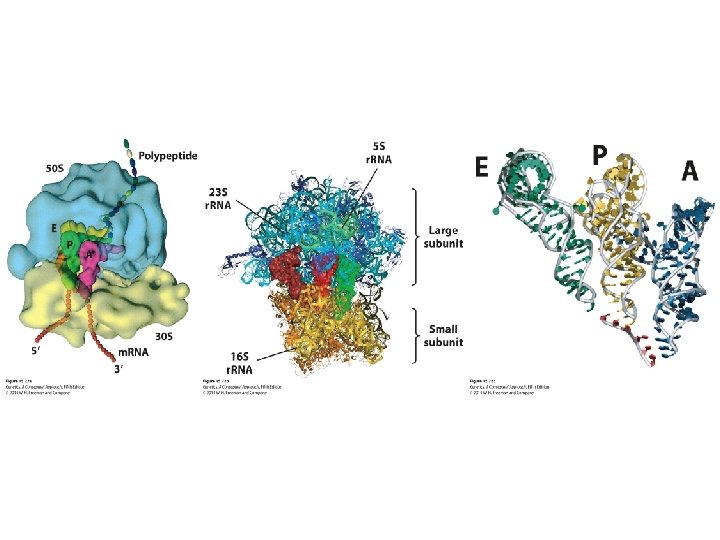
Elongation • Exit site E • Peptidyl site P • Aminoacyl site A • Elongation factors: Tu, Ts, and G
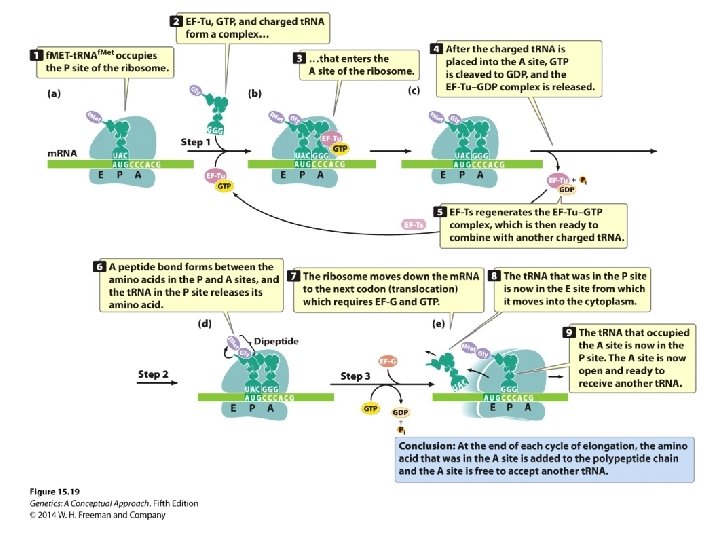
Concept Check 5 In elongation, the creation of peptide bonds between amino acids is catalyzed by. a. b. c. d. r. RNA protein in the small subunit protein in the large subunit t. RNA
Concept Check 5 In elongation, the creation of peptide bonds between amino acids is catalyzed by. a. b. c. d. r. RNA protein in the small subunit protein in the large subunit t. RNA
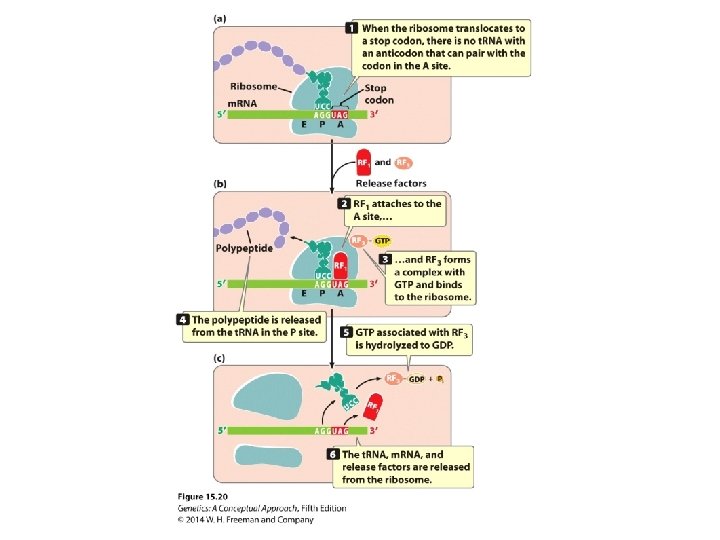
Termination • Termination codons: UAA, UAG, and UGA • Release factors
15. 4 Additional Properties of RNA and Ribosomes Affect Protein Synthesis • The three-dimensional structure of the ribosome • Polyribosomes: An m. RNA with several ribosomes attached
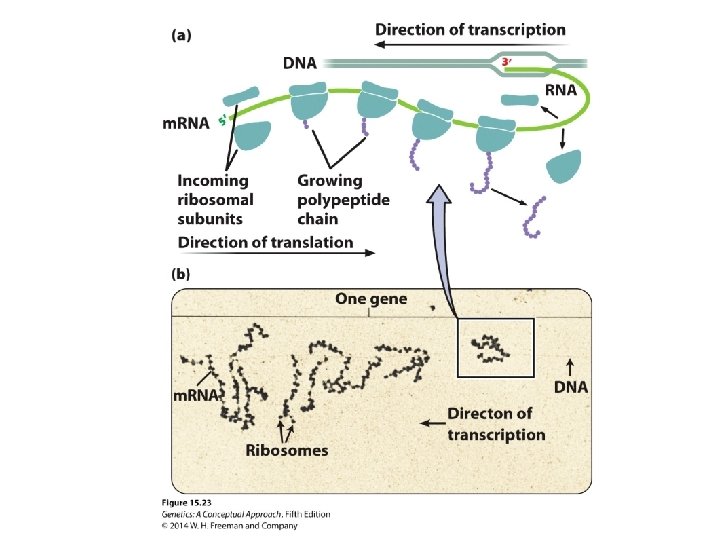
Concept Check 6 In a polyribosome, the polypeptides associated with which ribosomes will be the longest? a. b. c. d. Those at the 5′ end of m. RNA Those at the 3′ end of m. RNA Those in the middle of m. RNA All polypeptides will be the same length.
Concept Check 6 In a polyribosome, the polypeptides associated with which ribosomes will be the longest? a. b. c. d. Those at the 5′ end of m. RNA Those at the 3′ end of m. RNA Those in the middle of m. RNA All polypeptides will be the same length.
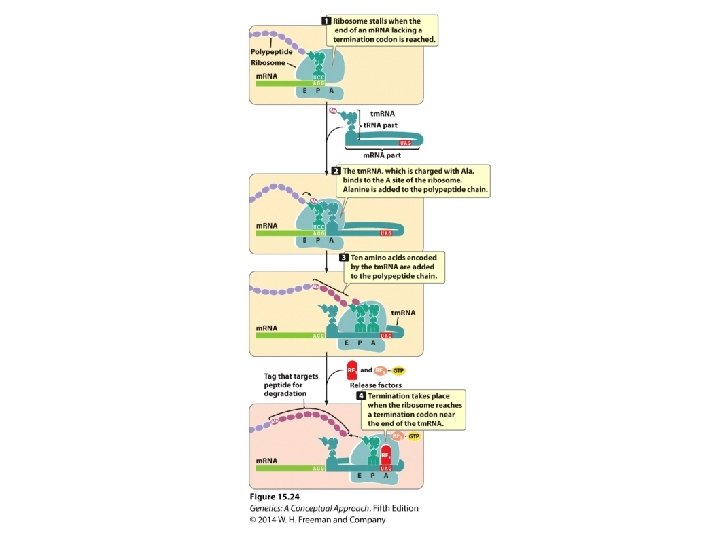
Additional Properties of RNA and Ribosomes Affect Protein Synthesis • Messenger RNA Surveillance • Detect and deal with errors in m. RNA • Nonsense–mediated m. RNA decay: eliminating m. RNA containing premature termination codons • Posttranslational Modifications of Proteins • Folding • Molecular Chaperones • Translation and Antiobiotics • Nonstandard Protein Synthesis
- Slides: 57