Bead Normalization of Mass Cytometry Data and MassTag
Bead Normalization of Mass Cytometry Data and Mass-Tag Cellular Barcoding Rachel Finck May 7, 2014
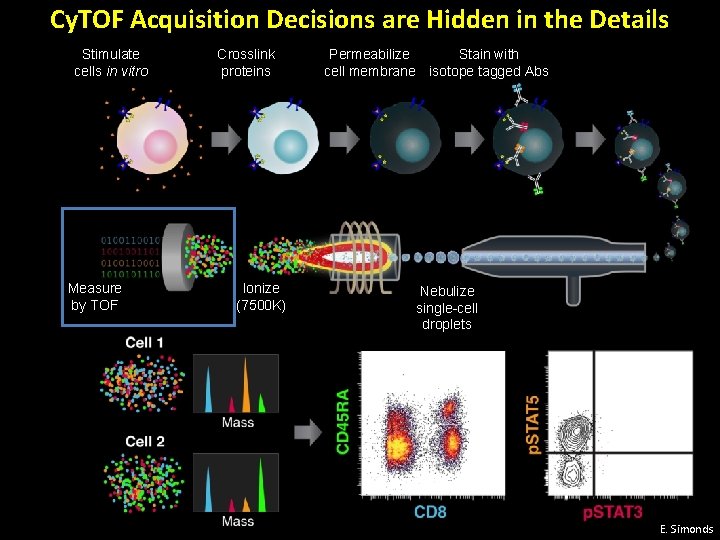
Cy. TOF Acquisition Decisions are Hidden in the Details Stimulate cells in vitro Measure by TOF Crosslink proteins Ionize (7500 K) Permeabilize Stain with cell membrane isotope tagged Abs Nebulize single-cell droplets E. Simonds
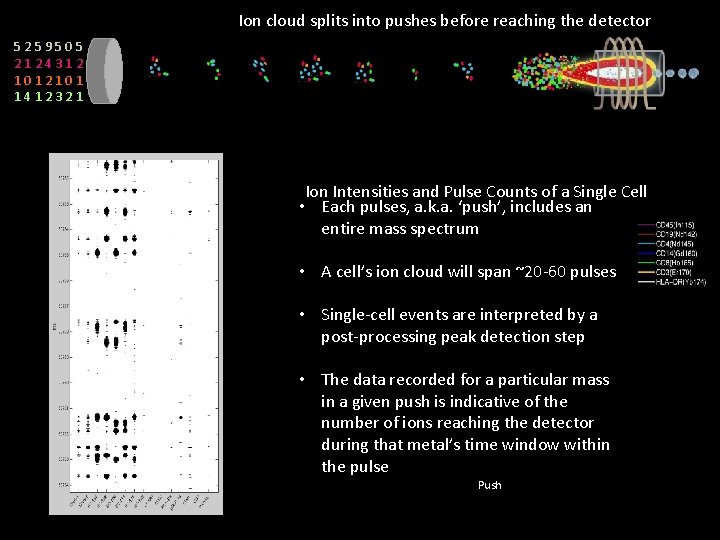
Ion cloud splits into pushes before reaching the detector 5259505 2124312 1012101 1412321 Ion Intensities and Pulse Counts of a Single Cell • Each pulses, a. k. a. ‘push’, includes an entire mass spectrum • A cell’s ion cloud will span ~20 -60 pulses • Single-cell events are interpreted by a post-processing peak detection step • The data recorded for a particular mass in a given push is indicative of the number of ions reaching the detector during that metal’s time window within the pulse Push

Ease of Quality Assessment/Control Can Be Obfuscated by High-Dimensional Analyses SPADE CD 3+ T cells CD 4+T cells Vi. SNE vs. CD 8+T cells Wanderlust

Ease of Quality Assessment/Control Can Be Obfuscated by High-Dimensional Analyses vs.
Outline – – Normalization of Mass Cytometry Data with Bead Standards • Review of normalization algorithm • Demonstration of normalization software with updated beads Mass-Tag Cellular Barcoding • Dose response to inhibitors • Single-cell deconvolution of barcoded populations • “Doublet-free” barcoding scheme
Normalization of Mass Cytometry Data Goal: Reliably compare mass cytometry data across patients, conditions, tissues, etc. Problem: Drifts in mass cytometry instrument sensitivity over time due to cellular debris, fluctuations in plasma temperature, and calibrations. Solution: Normalization using internal bead standards measured concurrently with cell samples.
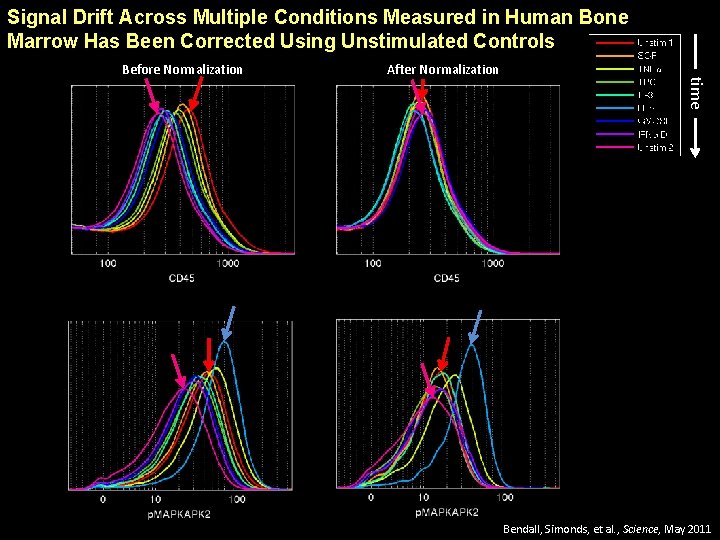
Signal Drift Across Multiple Conditions Measured in Human Bone Marrow Has Been Corrected Using Unstimulated Controls After Normalization time Before Normalization Bendall, Simonds, et al. , Science, May 2011
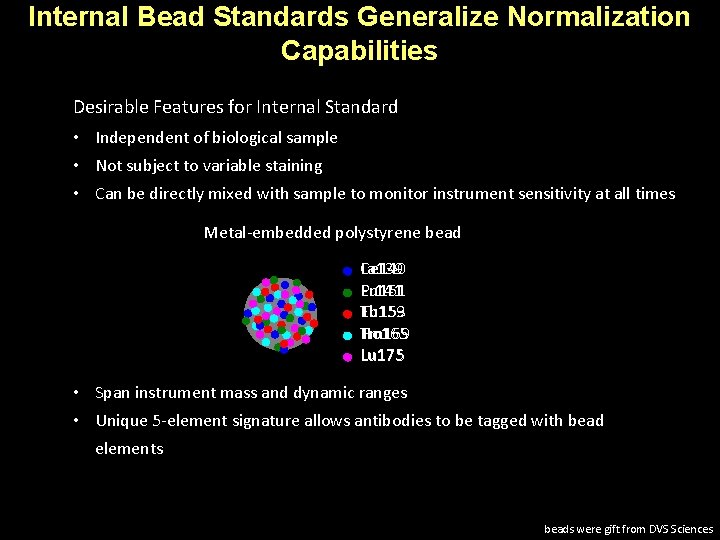
Internal Bead Standards Generalize Normalization Capabilities Desirable Features for Internal Standard • Independent of biological sample • Not subject to variable staining • Can be directly mixed with sample to monitor instrument sensitivity at all times Metal-embedded polystyrene bead La 139 Ce 140 Pr 141 Eu 151 Tb 159 Eu 153 Tm 169 Ho 165 Lu 175 • Span instrument mass and dynamic ranges • Unique 5 -element signature allows antibodies to be tagged with bead elements beads were gift from DVS Sciences

Beads can be spiked into standard mass cytometry protocol Stimulate cells in vitro Measure by TOF Crosslink proteins Ionize (7500 K) Permeabilize Stain with cell membrane isotope tagged Abs Nebulize single-cell droplets cells beads E. Simonds
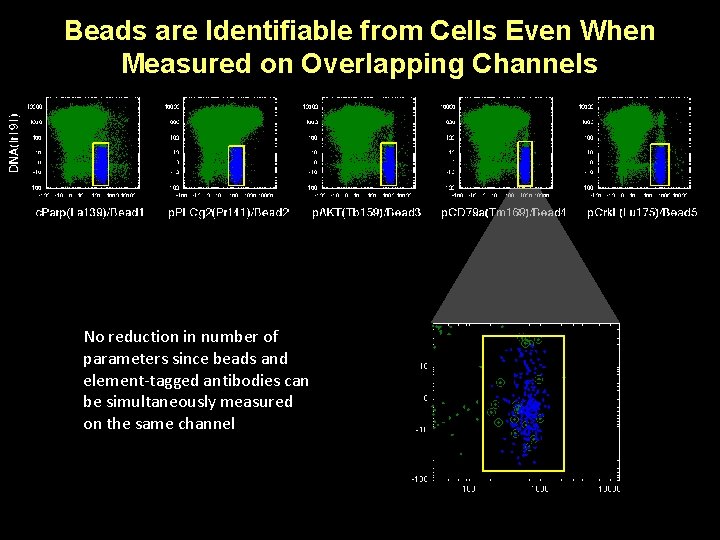
Beads are Identifiable from Cells Even When Measured on Overlapping Channels No reduction in number of parameters since beads and element-tagged antibodies can be simultaneously measured on the same channel
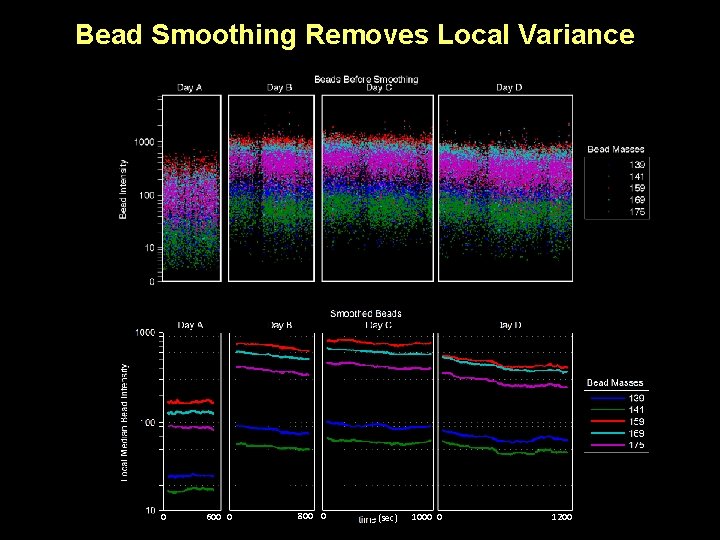
Bead Smoothing Removes Local Variance 0 600 0 800 0 (sec) 1000 0 1200
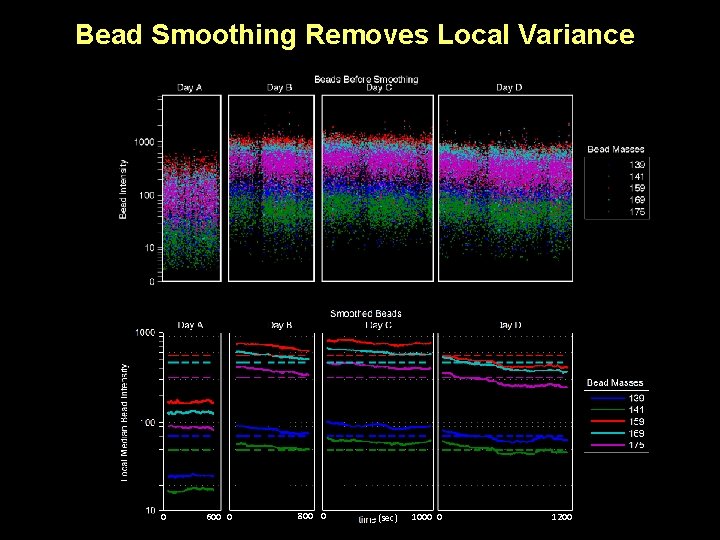
Bead Smoothing Removes Local Variance 0 600 0 800 0 (sec) 1000 0 1200
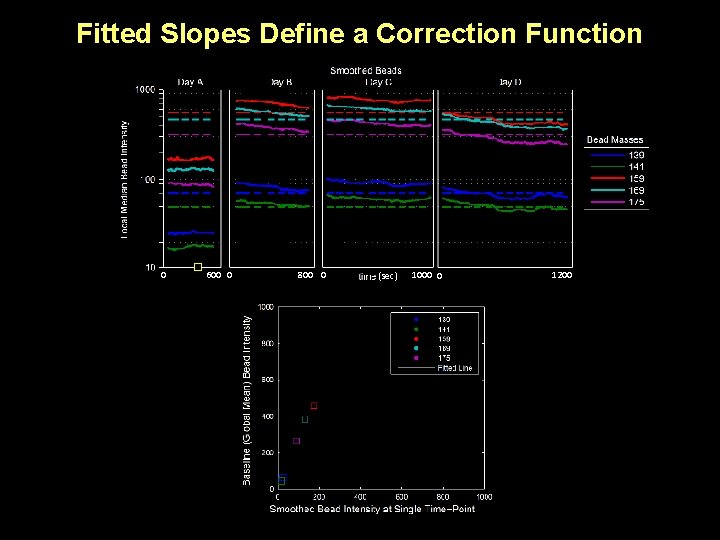
Fitted Slopes Define a Correction Function 0 600 0 800 0 (sec) 1000 0 1200
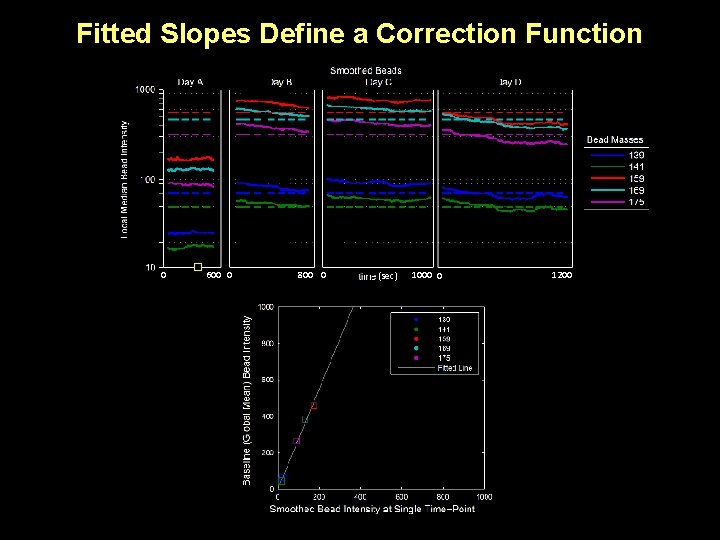
Fitted Slopes Define a Correction Function 0 600 0 800 0 (sec) 1000 0 1200
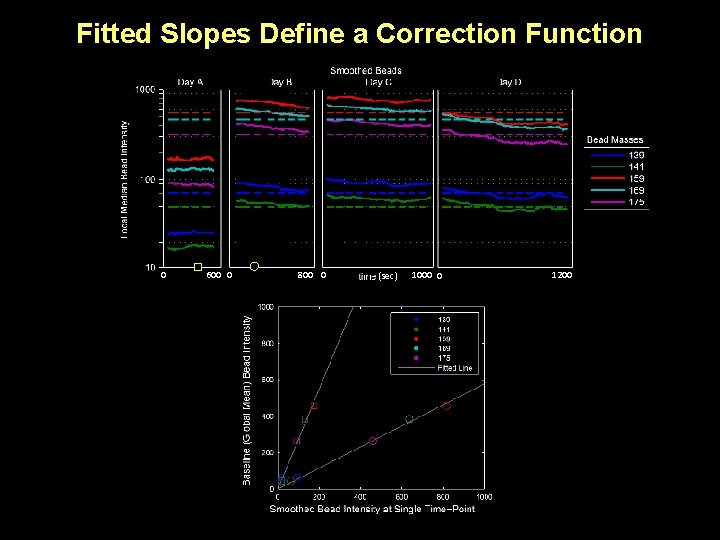
Fitted Slopes Define a Correction Function 0 600 0 800 0 (sec) 1000 0 1200
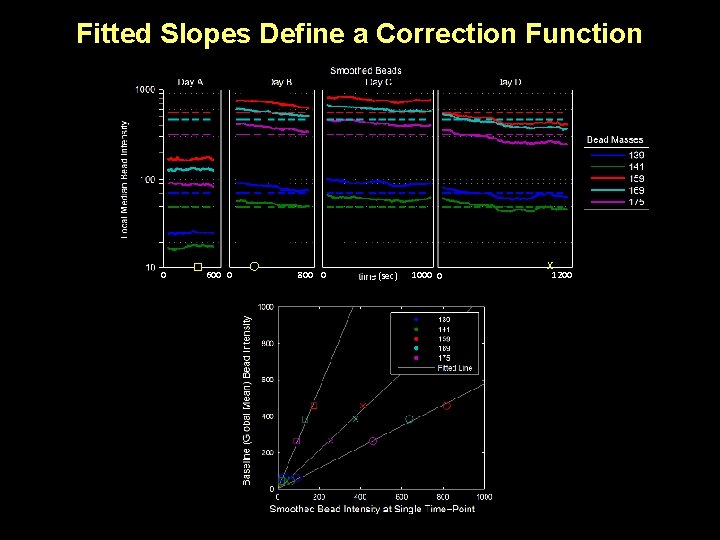
Fitted Slopes Define a Correction Function 0 600 0 800 0 (sec) 1000 0 x 1200

Fitted Slopes Define a Correction Function 0 600 0 800 0 (sec) 1000 0 1200

Normalization Reduces Bead Intensity Variation 4. 9 -fold mean range 1. 3 -fold mean range 0 600 0 800 0 (sec) 1000 0 1200
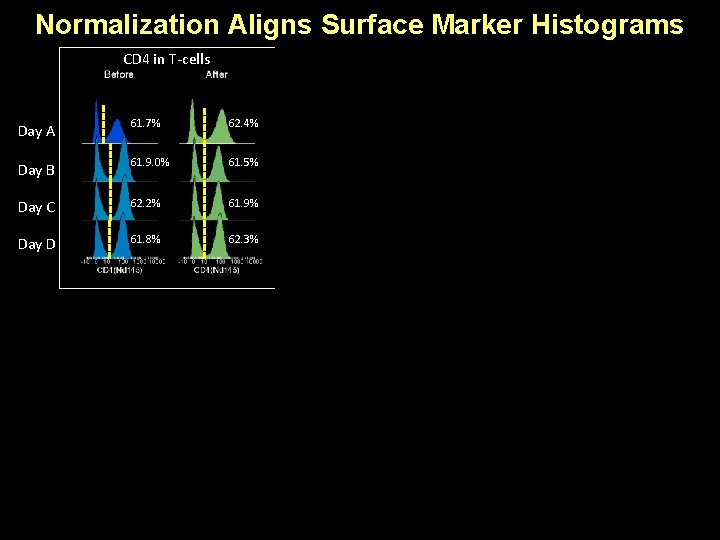
Normalization Aligns Surface Marker Histograms CD 4 in T-cells CD 8 in T-cells CD 19 in B-cells Day A 61. 7% 62. 4% 36. 5% 37. 2% 97. 1% 97. 3% Day B 61. 9. 0% 61. 5% 35. 8% 34. 8% 98. 0% 97. 8% Day C 62. 2% 61. 9% 35. 4% 34. 1% 97. 7% 97. 5% Day D 61. 8% 62. 3% 34. 7% 98. 2% 98. 1% CD 14 in Monocytes HLA-DR in Monocytes CD 38 in Monocytes Day A 87. 0% 87. 2% 92. 8% 93. 8% 97. 5% Day B 87. 8% 87. 4% 97. 3% 96. 7% 98. 5% Day C 89. 2% 88. 3% 97. 9% 97. 1% 98. 7% 98. 5% Day D 88. 8% 89. 4% 97. 6% 97. 2% 98. 6%
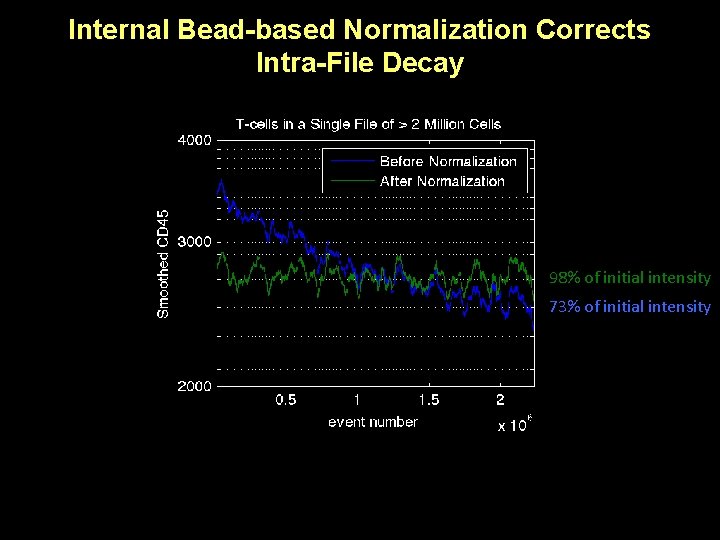
Internal Bead-based Normalization Corrects Intra-File Decay 98% of initial intensity 73% of initial intensity
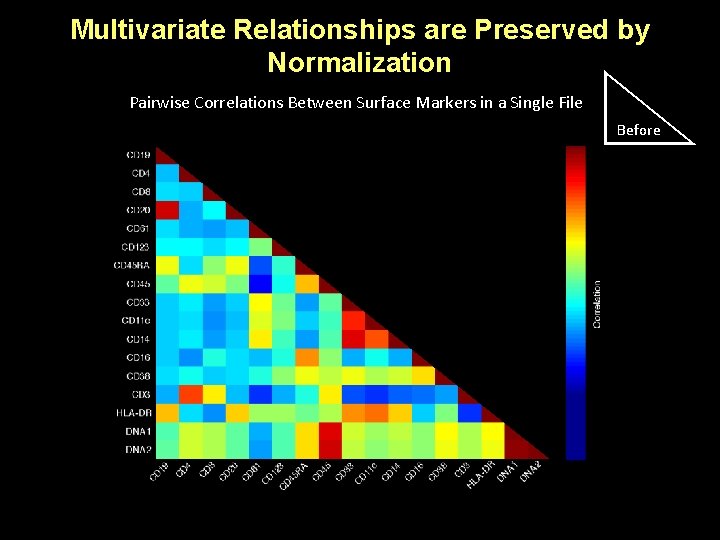
Multivariate Relationships are Preserved by Normalization Pairwise Correlations Between Surface Markers in a Single File After Before

Normalization Reduced Non-Biological Variation in Primary Pediatric ALL Bone Marrow Samples 4. 1 -fold mean range 1. 2 -fold mean range

patient Normalization Reduced Non-Biological Variation in Primary Pediatric ALL Bone Marrow Samples
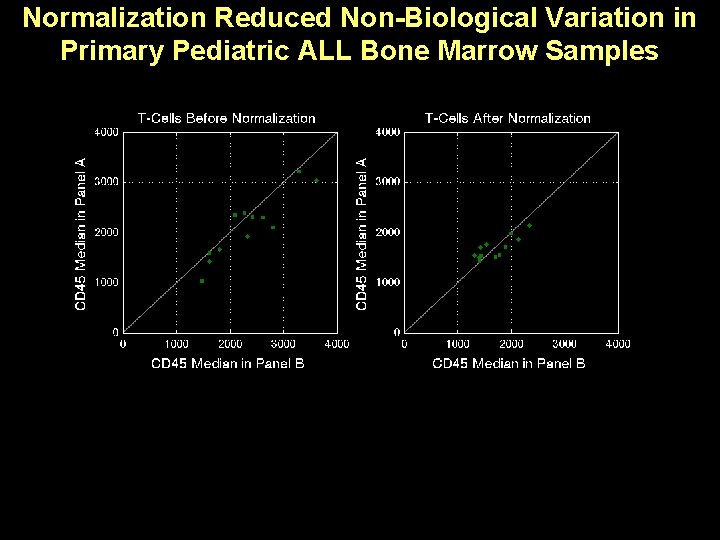
Normalization Reduced Non-Biological Variation in Primary Pediatric ALL Bone Marrow Samples

Normalization Software Example Using New Beads 1. Adjust gates. 2. Approve gates. 3. Set bead removal threshold.
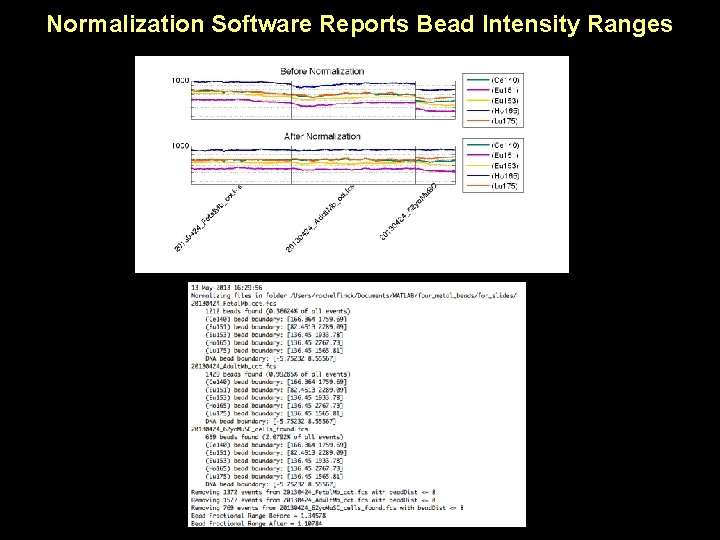
Normalization Software Reports Bead Intensity Ranges
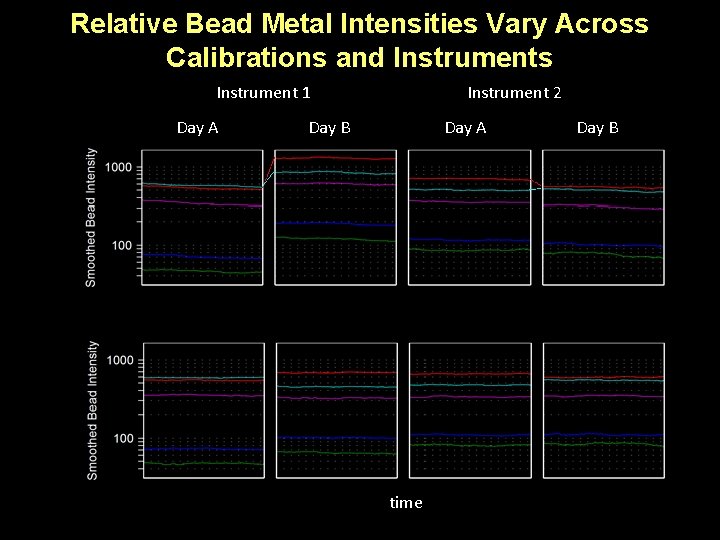
Relative Bead Metal Intensities Vary Across Calibrations and Instruments Instrument 1 Day A Instrument 2 Day B Day A time Day B
Normalization Summary • the intensities of the normalization beads quantitatively monitor instrument sensitivity over acquisition time • the effects of instrument variation on mass cytometry data are reduced by a correction derived from internal 5 metal bead standards • normalization allows for the direct comparison of samples collected from multiple individuals and treated under different conditions

Bernd Bodenmiller Eli Zunder Multiplexed mass cytometry profiling of cellular states perturbed by smallmolecule regulators Bernd Bodenmiller*, Eli R. Zunder*, Rachel Finck*, Tiffany J. Chen, Erica S. Savig, Robert V. Bruggner, Erin F. Simonds, Sean C. Bendall, Karen Sachs, Peter O. Krutzik and Garry P. Nolan Nature Biotechnology. 2012 September 10; 30(9): 858 -67
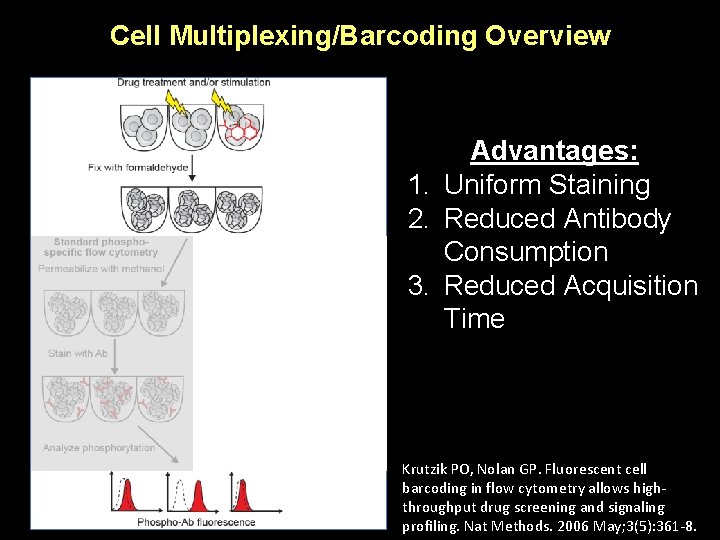
Cell Multiplexing/Barcoding Overview 1. 2. 3. 4. Advantages: Uniform Staining Reduced Antibody Consumption Reduced Acquisition Time Improved Singlet Detection Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows highthroughput drug screening and signaling profiling. Nat Methods. 2006 May; 3(5): 361 -8.
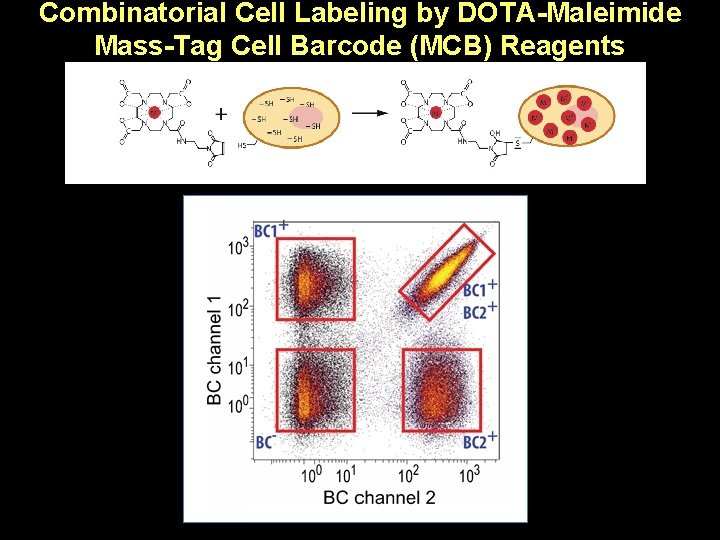
Combinatorial Cell Labeling by DOTA-Maleimide Mass-Tag Cell Barcode (MCB) Reagents
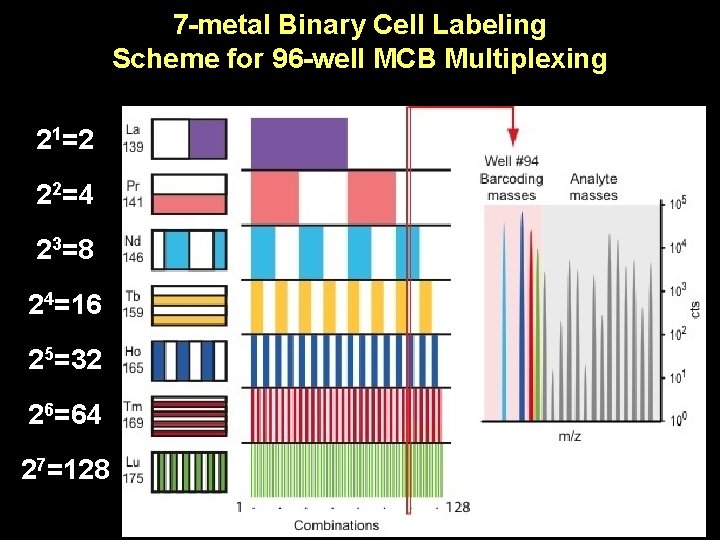
7 -metal Binary Cell Labeling Scheme for 96 -well MCB Multiplexing 21=2 22=4 23=8 24=16 25=32 26=64 27=128
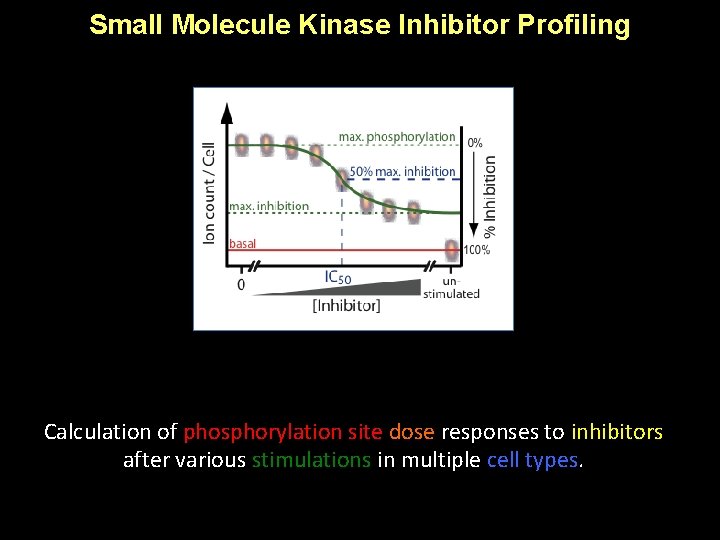
Small Molecule Kinase Inhibitor Profiling Calculation of phosphorylation site dose responses to inhibitors after various stimulations in multiple cell types.

Small Molecule Kinase Inhibitor Profiling
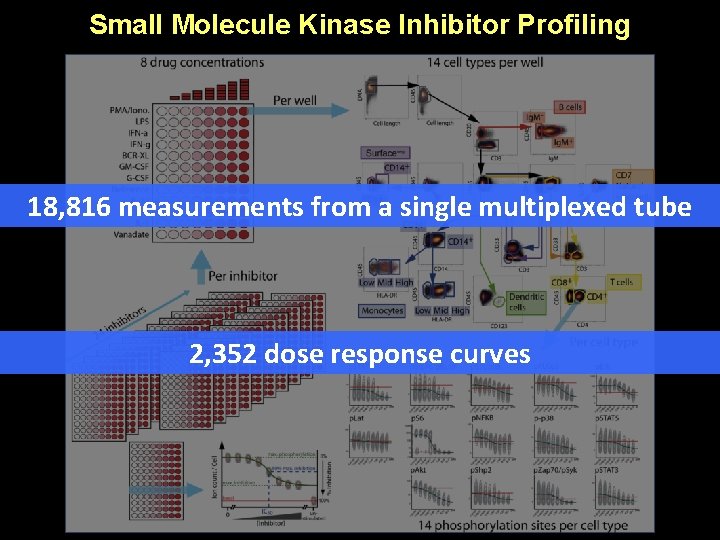
Small Molecule Kinase Inhibitor Profiling 18, 816 measurements from a single multiplexed tube 2, 352 dose response curves
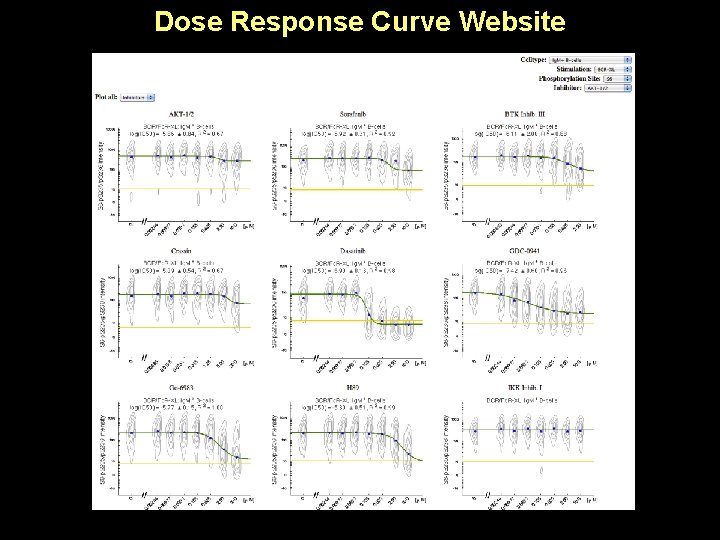
Dose Response Curve Website
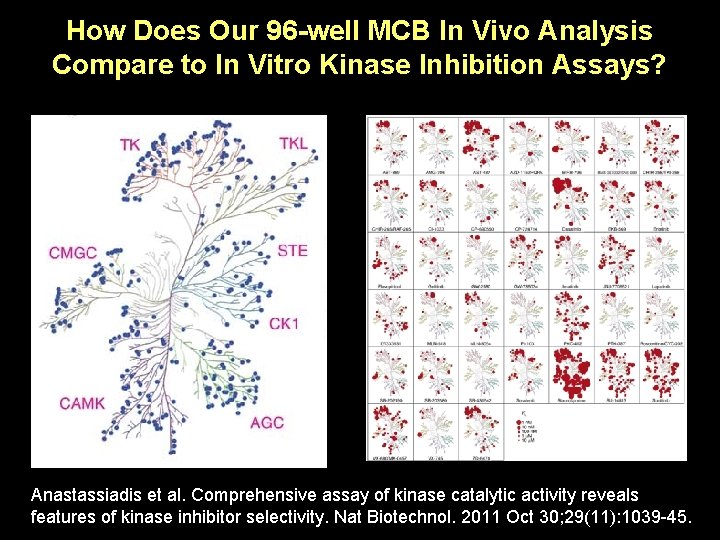
How Does Our 96 -well MCB In Vivo Analysis Compare to In Vitro Kinase Inhibition Assays? Anastassiadis et al. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol. 2011 Oct 30; 29(11): 1039 -45.

PBMC In Vivo IC 50 s Strongly Correlate with In Vitro Percent Inhibition Values Anastassiadis et al. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol. 2011 Oct 30; 29(11): 1039 -45. Davis et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011 Oct 30; 29(11): 1046 -51.

Prof. Garry P. Nolan www. stanford. edu/group/nolan Sean Bendall Erin Simonds Astraea Jager Kara Davis Wendy Fantl Eli Zunder Bernd Bodenmiller Rob Bruggner Karen Sachs Greg Behbehani Columbia Dana Pe’er Smita Krishnaswamy El-ad David Amir U. Toronto / DVS Sciences Scott Tanner Olga Ornatsky Vladimir Baranov Dmitry Bandura Tad George
- Slides: 40