Basic Features of Eukaryotic Genes Genetic Markers RFLP

Basic Features of Eukaryotic Genes Genetic Markers: RFLP & LOH G. Mathan Ph. D. , Assistant Professor
Non coding DNA • In genetics, noncoding DNA describes components of an organism's DNA sequences that do not encode for protein sequences. • In many eukaryotes, a large percentage of an organism's total genome size is noncoding DNA
Types of non coding DNA sequence § Non coding RNA: functional RNA molecules that are not translated into protein. Examples ribosomal RNA, t RNA and micro RNA. § § Cis- and Trans-regulatory elements: Introns Pseudogenes Telomeres
Genetic Markers A genetic marker is a gene or DNA sequence with a known location on a chromosome that can be used to identify cells, individuals or species
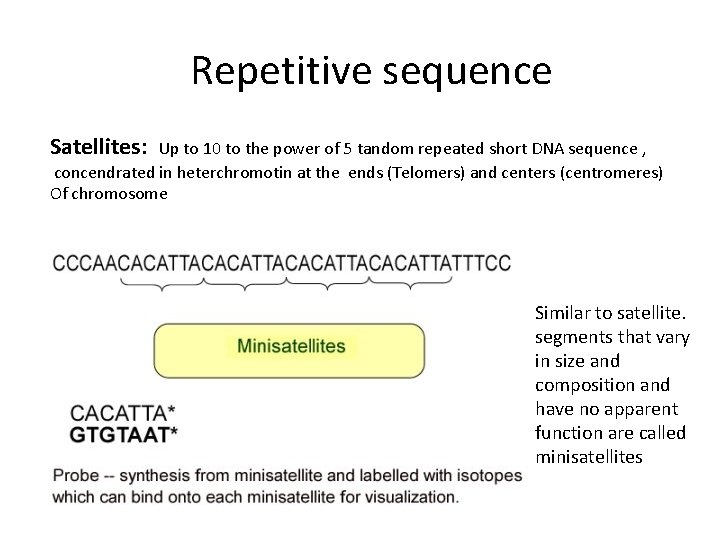
Repetitive sequence Satellites: Up to 10 to the power of 5 tandom repeated short DNA sequence , concendrated in heterchromotin at the ends (Telomers) and centers (centromeres) Of chromosome Similar to satellite. segments that vary in size and composition and have no apparent function are called minisatellites

How do we distinguish one person’s DNA from another? We do not need to sequence the entire 3 billion base pairs of a person’s DNA to distinguish it from another person’s DNA u Intron regions of DNA (junk DNA) contain sequences that are 20 -100 bp in length that are repeated at different locations (loci) along the chromosome. CGGCTACGGCTA (repeated 3 times at this location; at another location, it may be repeated 9 times) u These sequences are called Short Tandem Repeats (STRs) or VNTRs u
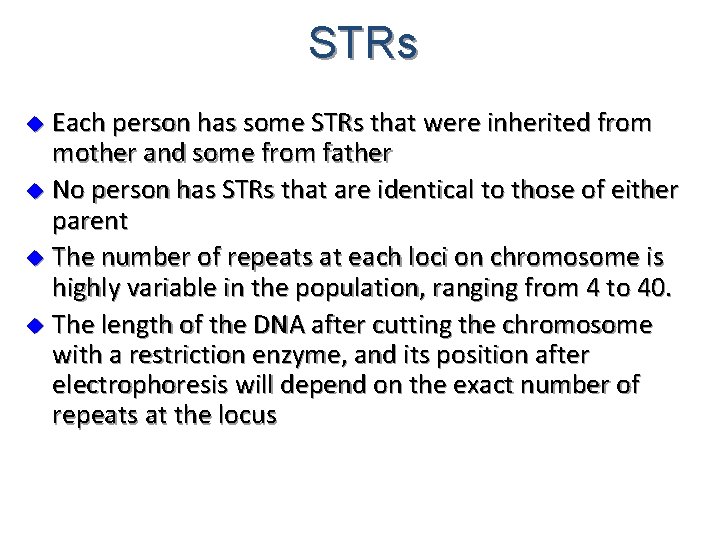
STRs Each person has some STRs that were inherited from mother and some from father u No person has STRs that are identical to those of either parent u The number of repeats at each loci on chromosome is highly variable in the population, ranging from 4 to 40. u The length of the DNA after cutting the chromosome with a restriction enzyme, and its position after electrophoresis will depend on the exact number of repeats at the locus u

u The uniqueness of an individual’s STRs provides the scientific marker of identity known as a DNA fingerprint. u In the United States the FBI has standardized a set of 13 STR assays (13 different locations on the chromosomes) for DNA typing, and has organized the CODIS database forensic identification in criminal cases. u The United States maintains the largest DNA database in the world: The Combined DNA Index System, with over 60 million records as of 2007.
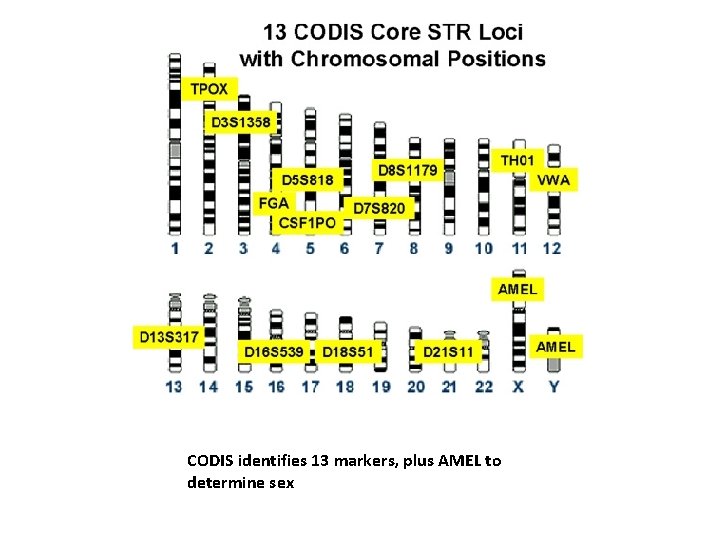
CODIS identifies 13 markers, plus AMEL to determine sex
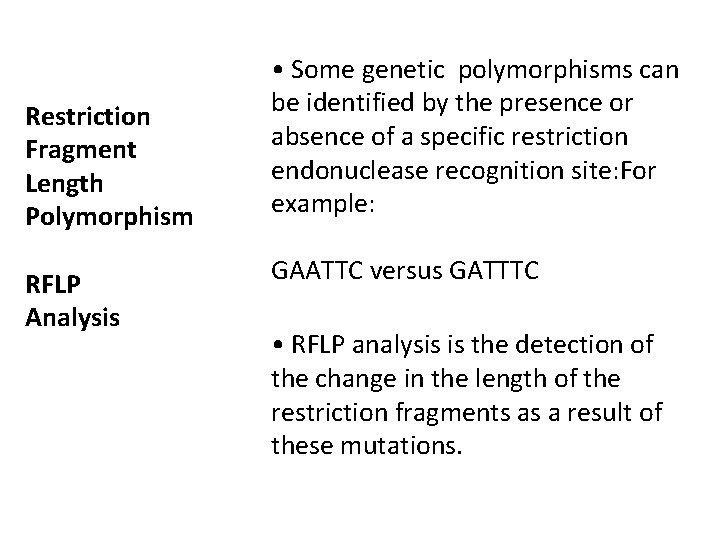
Restriction Fragment Length Polymorphism RFLP Analysis • Some genetic polymorphisms can be identified by the presence or absence of a specific restriction endonuclease recognition site: For example: GAATTC versus GATTTC • RFLP analysis is the detection of the change in the length of the restriction fragments as a result of these mutations.

DNA Profiling • A technique used by scientists to distinguish between individuals of the same species using only samples of their DNA • The process of DNA fingerprinting was invented by Alec Jeffreys at the University of Leicester in 1985.
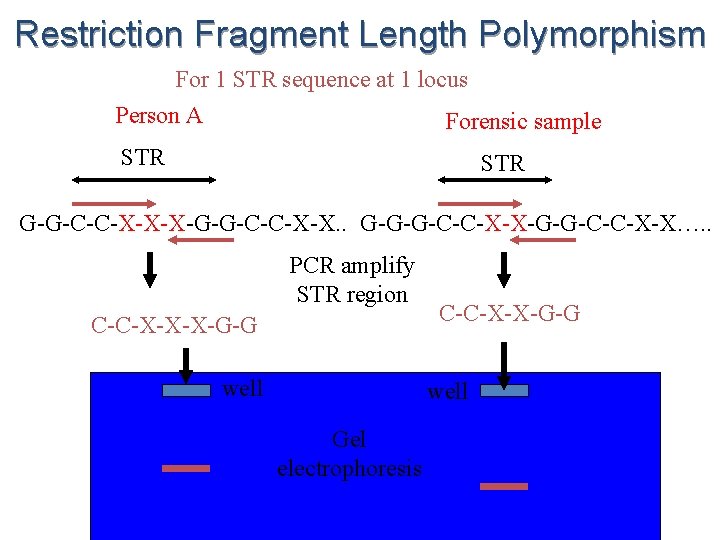
Restriction Fragment Length Polymorphism For 1 STR sequence at 1 locus Person A Forensic sample STR G-G-C-C-X-X-X-G-G-C-C-X-X. . G-G-G-C-C-X-X…. . PCR amplify STR region C-C-X-X-X-G-G well C-C-X-X-G-G well Gel electrophoresis

u If you do this for 13 different repeat sequences at 13 different loci on the chromosome, each person produces a different band pattern when the fragments are separated by gel electrophoresis
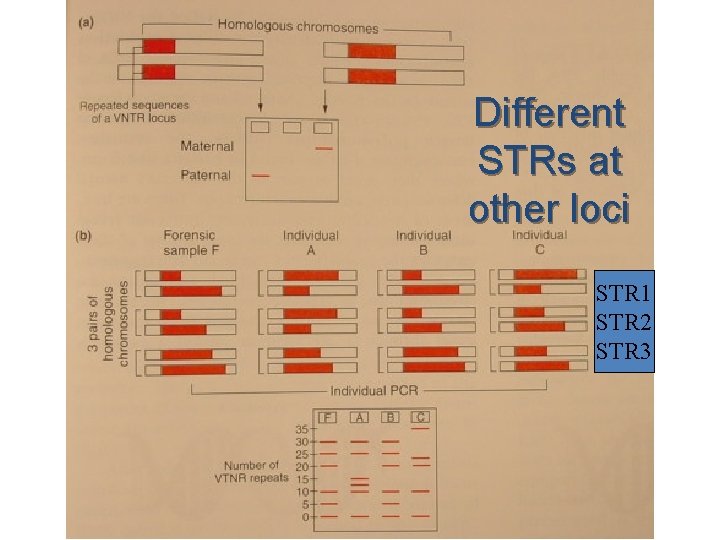
Different STRs at other loci STR 1 STR 2 STR 3

Chain of custody u Requires that collection of evidence must be systematically recorded and access to evidence must be controlled – Special challenges for DNA samples u crime scene may have DNA from people other than perpetrators of crime. These people could become suspects based on this DNA u DNA collected from victims in a morgue can become contaminated by DNA of other bodies previously on autopsy table u during early days all procedures for processing DNA was not standardized, people running assays were not experienced and made mistakes
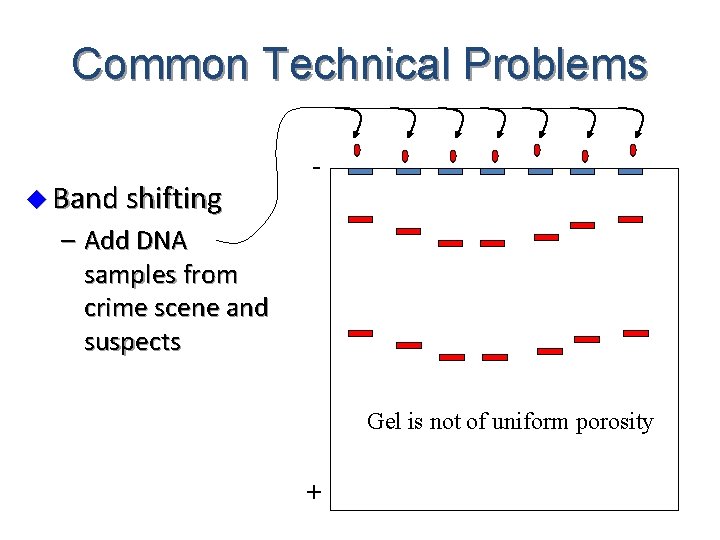
Common Technical Problems u Band shifting - – Add DNA samples from crime scene and suspects Gel is not of uniform porosity +
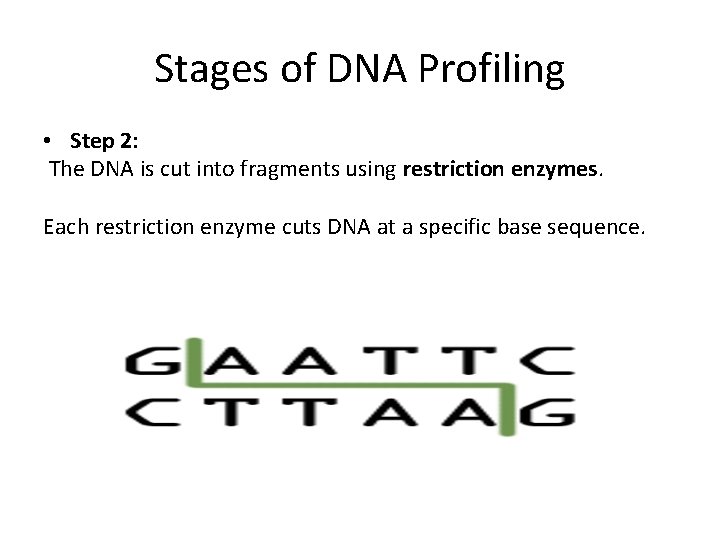
Stages of DNA Profiling • Step 2: The DNA is cut into fragments using restriction enzymes. Each restriction enzyme cuts DNA at a specific base sequence.
Stages of DNA Profiling • The sections of DNA that are cut out are called restriction fragments. • This yields thousands of restriction fragments of all different sizes because the base sequences being cut may be far apart (long fragment) or close together (short fragment).
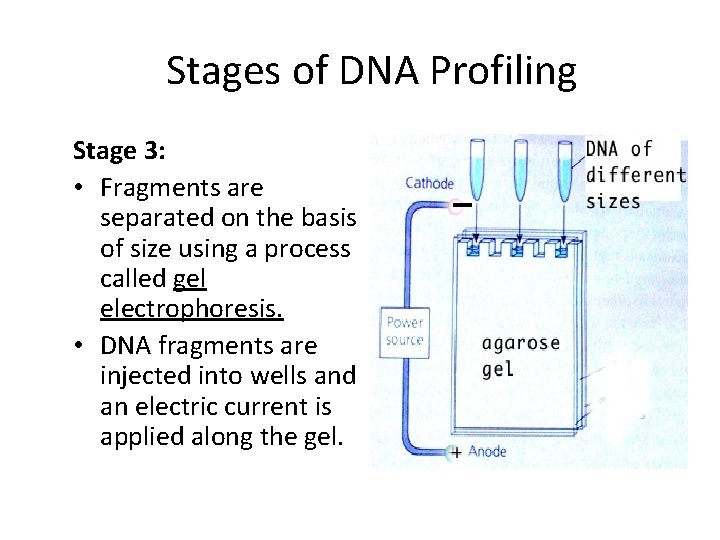
Stages of DNA Profiling Stage 3: • Fragments are separated on the basis of size using a process called gel electrophoresis. • DNA fragments are injected into wells and an electric current is applied along the gel.
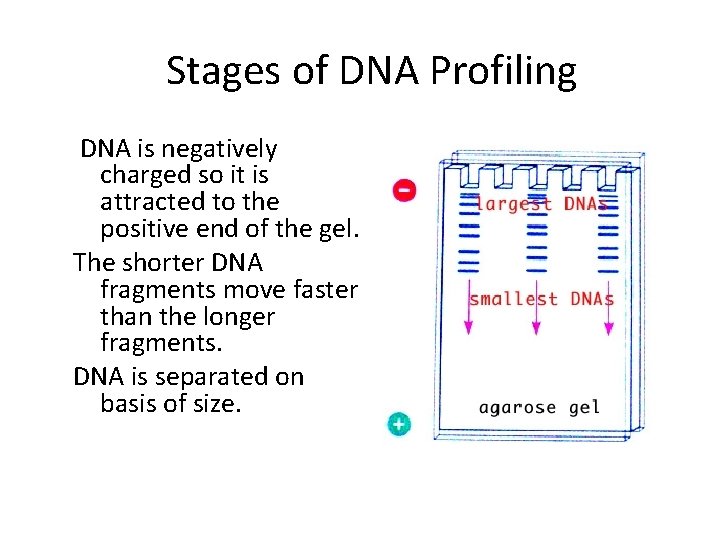
Stages of DNA Profiling DNA is negatively charged so it is attracted to the positive end of the gel. The shorter DNA fragments move faster than the longer fragments. DNA is separated on basis of size.
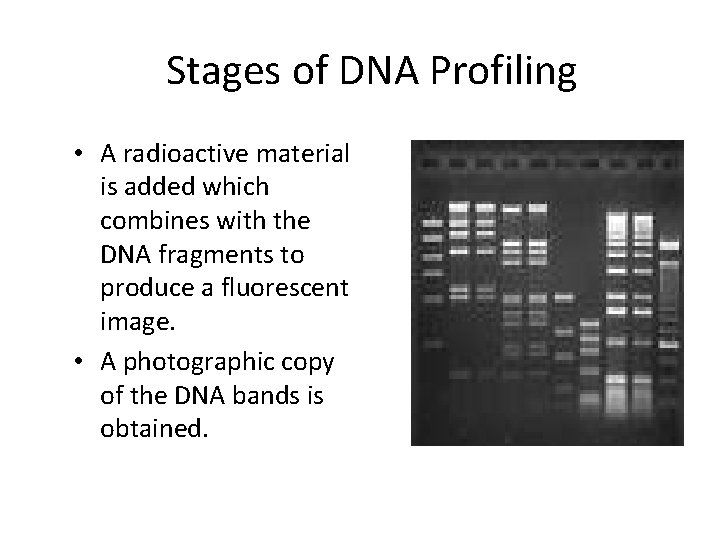
Stages of DNA Profiling • A radioactive material is added which combines with the DNA fragments to produce a fluorescent image. • A photographic copy of the DNA bands is obtained.
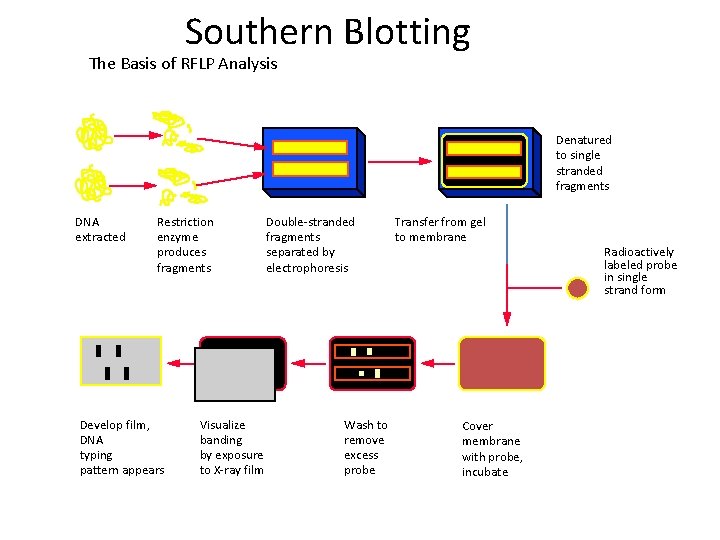
Southern Blotting The Basis of RFLP Analysis Denatured to single stranded fragments DNA extracted Restriction enzyme produces fragments Develop film, DNA typing pattern appears Visualize banding by exposure to X-ray film Double-stranded fragments separated by electrophoresis Wash to remove excess probe Transfer from gel to membrane Cover membrane with probe, incubate Radioactively labeled probe in single strand form
Stages of DNA Profiling Stage 4: • The pattern of fragment distribution is then analysed. • DNA profiling is used to solve crimes and medical problems
DNA profiling of ROMANOVs Bone 1990 S – DNA profile in a kinship study- on the bones of Romanovs Last member of Russia in ruling family. Russian revolution – Tsar Nicholas and his wife Tsarina Alexandria and their 5 children's were imprisoned -In 1917 all the 7 were killed along with their doctor and servants. In 1991 after the fall of communism the bodies were recovered from road side grave. STR ANALYSIS : Body recovered – more than a collection of bones. Mixed up Mitochondrial DNA analysis- TO CONFIRM THEIR BONES ARE AUTHENTICATED.
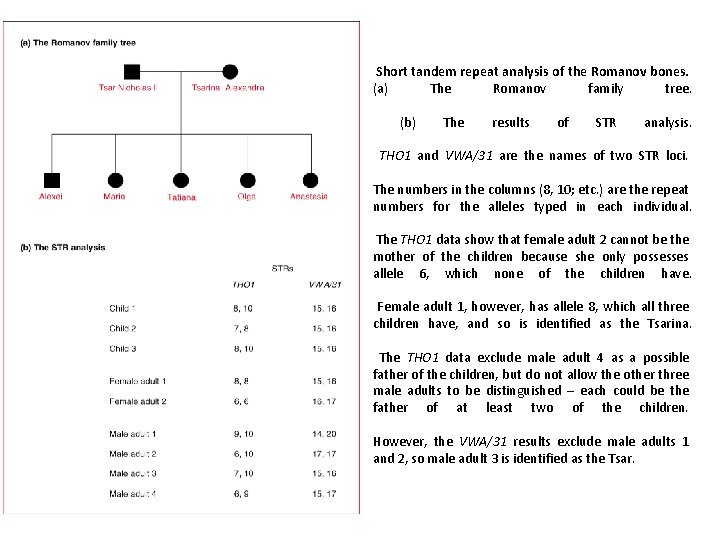
Short tandem repeat analysis of the Romanov bones. (a) The Romanov family tree. (b) The results of STR analysis. THO 1 and VWA/31 are the names of two STR loci. The numbers in the columns (8, 10; etc. ) are the repeat numbers for the alleles typed in each individual. The THO 1 data show that female adult 2 cannot be the mother of the children because she only possesses allele 6, which none of the children have. Female adult 1, however, has allele 8, which all three children have, and so is identified as the Tsarina. The THO 1 data exclude male adult 4 as a possible father of the children, but do not allow the other three male adults to be distinguished – each could be the father of at least two of the children. However, the VWA/31 results exclude male adults 1 and 2, so male adult 3 is identified as the Tsar.
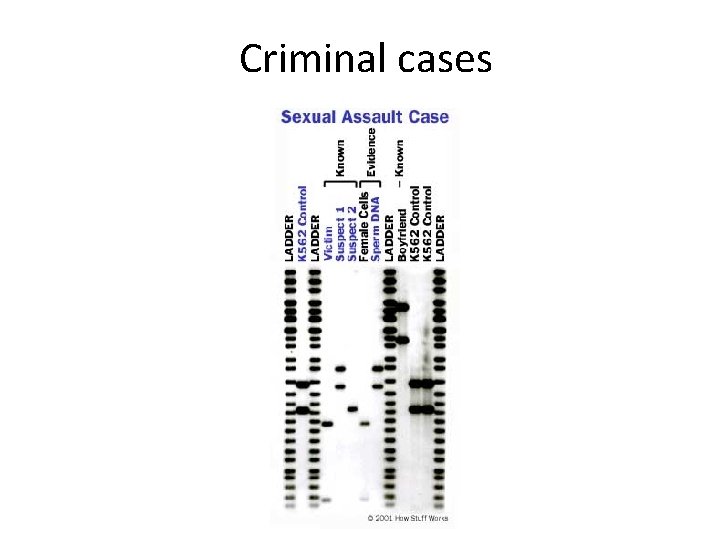
Criminal cases
What is a SNP? • Mutation of a single nucleotide (A, C, T, G) • Some can be associated with various phenotypic differences – Drug resistance – Propensity towards disease • Over 5 Million SNP locations identified in the human genome, growing daily

SNPs and Cancer • Tumors frequently exhibit DNA alterations – Loss (0 or 1 copy) – Additions (3 or more copies) • Could be responsible for some cancers – Tumor suppresor genes – Oncogenes
Loss-of-Heterozygosity (LOH) • Loss of heterozygosity (LOH) in a cell is the loss of normal function of one allele of a gene in which the other allele was already inactivated. • If a marker (SNP, micro-satellite) has heterozygous genotype in the normal sample but has homozygous genotype in the tumor sample from the same patient. • Indicates chromosomal alteration; often related to tumor-suppressor genes
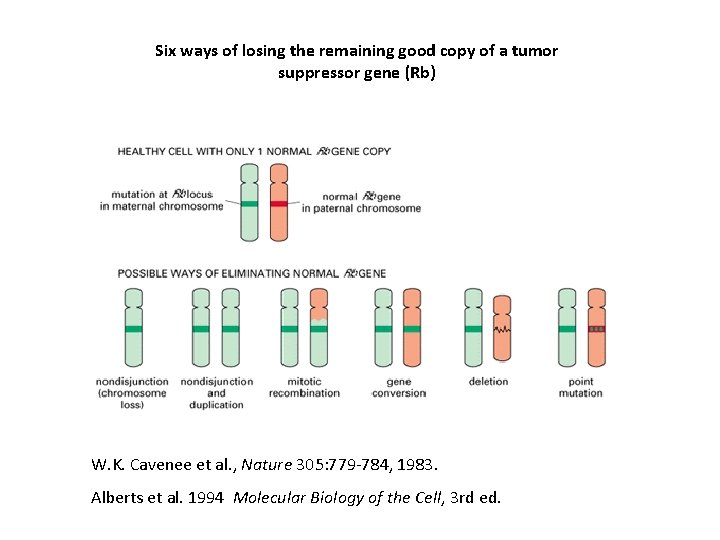
Six ways of losing the remaining good copy of a tumor suppressor gene (Rb) W. K. Cavenee et al. , Nature 305: 779 -784, 1983. Alberts et al. 1994 Molecular Biology of the Cell, 3 rd ed.

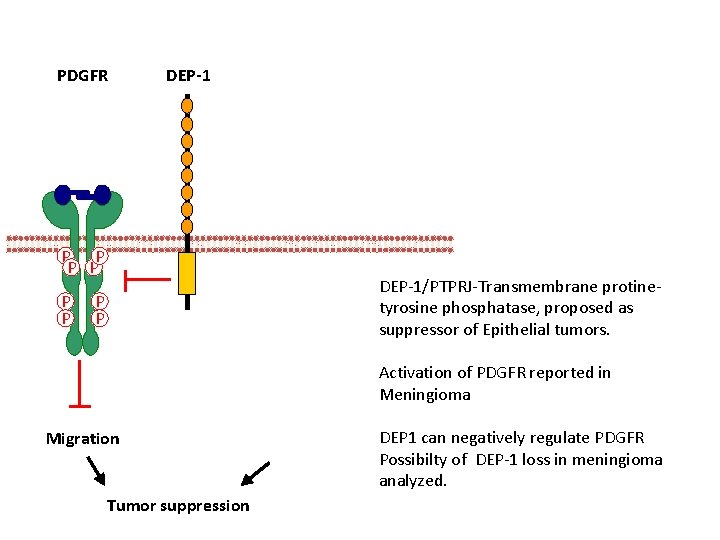
PDGFR DEP-1 P P P DEP-1/PTPRJ-Transmembrane protinetyrosine phosphatase, proposed as suppressor of Epithelial tumors. P P Activation of PDGFR reported in Meningioma Migration Tumor suppression DEP 1 can negatively regulate PDGFR Possibilty of DEP-1 loss in meningioma analyzed.
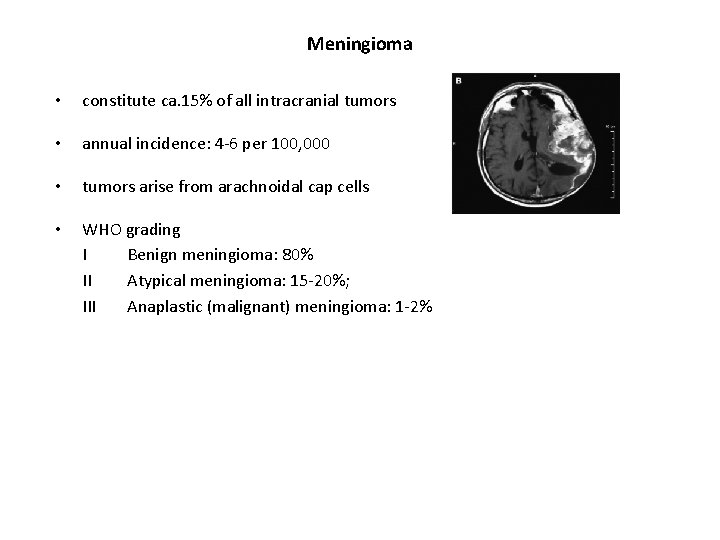
Meningioma • constitute ca. 15% of all intracranial tumors • annual incidence: 4 -6 per 100, 000 • tumors arise from arachnoidal cap cells • WHO grading I Benign meningioma: 80% II Atypical meningioma: 15 -20%; III Anaplastic (malignant) meningioma: 1 -2%
LOH analysis A total 32 different histological subtype and grade(WHO classification of brain tumors) of malignancy were used for the LOH analysis. DNA extraction from snap-frozen tumor samples DNA samples were amplified by PCR using fluorescently (Cy 5) labelled primers. PTPRJ- LOH analyzed at 4 microsatellite: D 11 S 1350 D 11 S 1784 D 11 S 4117 D 11 S 4183 NF 2 - 3 microsatellite markers D 22 S 268 D 22 S 421 D 22 S 929 A difference of 30% or more in the intensity ratio of the two alleles in tumor DNA compared to normal DNA was considered as evidence for LOH.
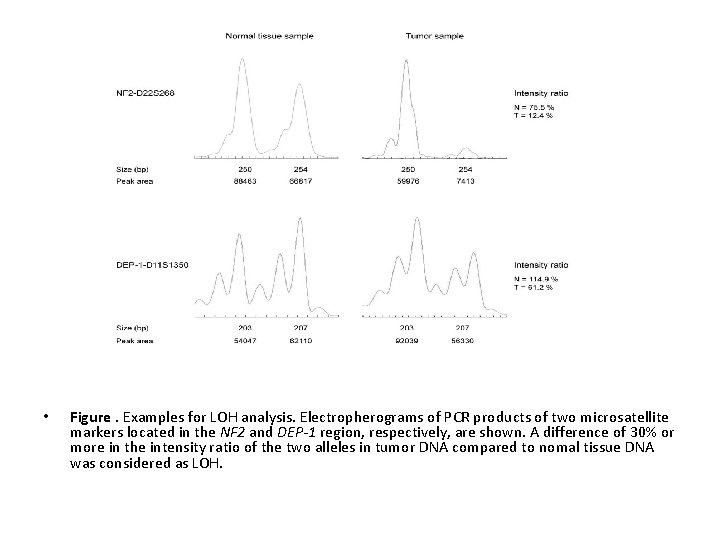
• Figure. Examples for LOH analysis. Electropherograms of PCR products of two microsatellite markers located in the NF 2 and DEP-1 region, respectively, are shown. A difference of 30% or more in the intensity ratio of the two alleles in tumor DNA compared to nomal tissue DNA was considered as LOH.
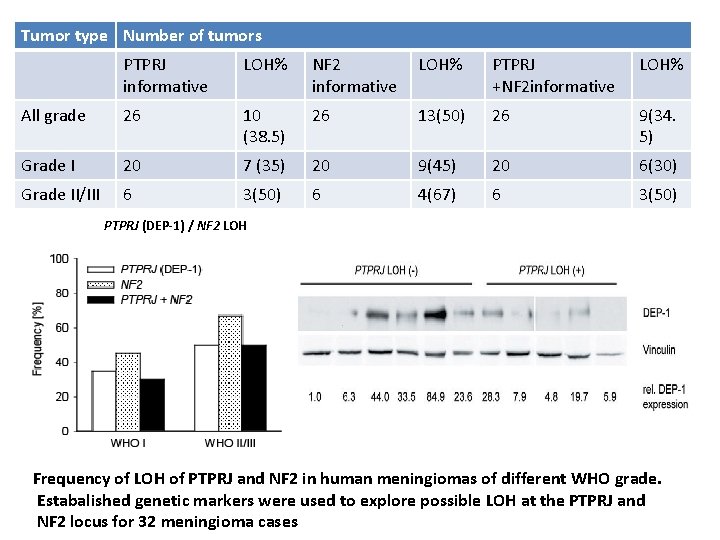
Tumor type Number of tumors PTPRJ informative LOH% NF 2 informative LOH% PTPRJ +NF 2 informative LOH% All grade 26 10 (38. 5) 26 13(50) 26 9(34. 5) Grade I 20 7 (35) 20 9(45) 20 6(30) Grade II/III 6 3(50) 6 4(67) 6 3(50) PTPRJ (DEP-1) / NF 2 LOH Frequency of LOH of PTPRJ and NF 2 in human meningiomas of different WHO grade. Estabalished genetic markers were used to explore possible LOH at the PTPRJ and NF 2 locus for 32 meningioma cases

Study Results • PTPRJ allelic loss and loss of DEP-1 protein expression occur in significant fraction of meningiomas , potentially indicating tumor biology

THANKS FOR YOUR ATTENTION!

Acknowledgement v The Presentation is being used for educational and non commercial purpose v Thanks are due to all those original contributors and entities whose pictures used for making this presentation.
- Slides: 39