Base quality and read quality How should data

Base quality and read quality: How should data quality be measured? Gabor T. Marth Boston College Biology Department 1000 Genomes Meeting Cold Spring Harbor Laboratory May 5 -6. 2008
Read accuracy / quality values Read accuracy • the more accurate the read, the easier and faster it is to map • the more errors the aligner must tolerate, the less reads can be uniquely aligned Well-calibrated quality values • quality values help distinguish between sequencing error and allelic difference • some aligners (e. g. MAQ) use quality values to find correct read mapping position
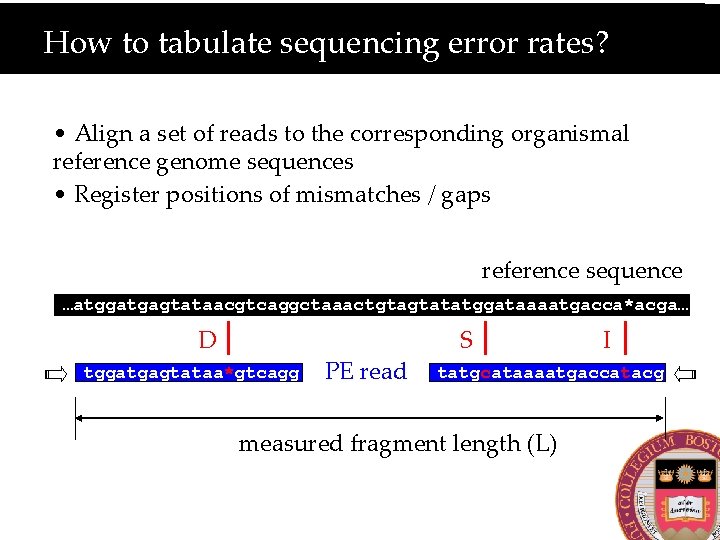
How to tabulate sequencing error rates? • Align a set of reads to the corresponding organismal reference genome sequences • Register positions of mismatches / gaps reference sequence …atggatgagtataacgtcaggctaaactgtagtatatggataaaatgacca*acga… D tggatgagtataa*gtcagg PE read S I tatgcataaaatgaccatacg measured fragment length (L)
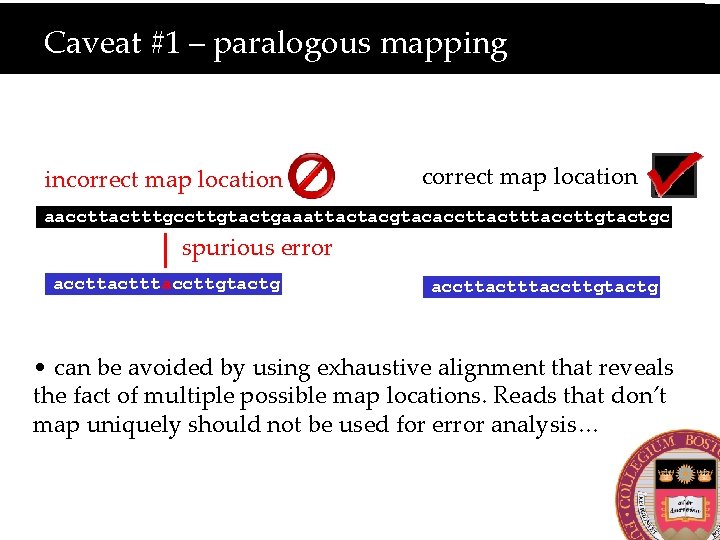
Caveat #1 – paralogous mapping incorrect map location aaccttactttgccttgtactgaaattactacgtacaccttactttaccttgtactgc spurious error accttactttaccttgtactg • can be avoided by using exhaustive alignment that reveals the fact of multiple possible map locations. Reads that don’t map uniquely should not be used for error analysis…
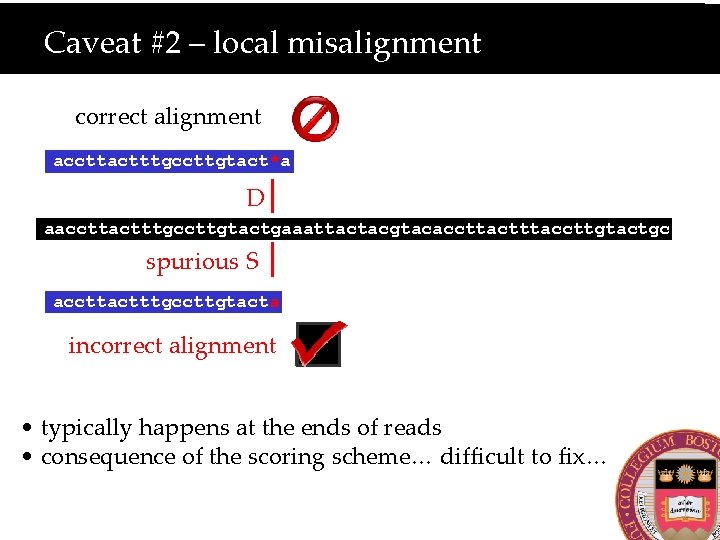
Caveat #2 – local misalignment correct alignment accttactttgccttgtact*a D aaccttactttgccttgtactgaaattactacgtacaccttactttaccttgtactgc spurious S accttactttgccttgtacta incorrect alignment • typically happens at the ends of reads • consequence of the scoring scheme… difficult to fix…
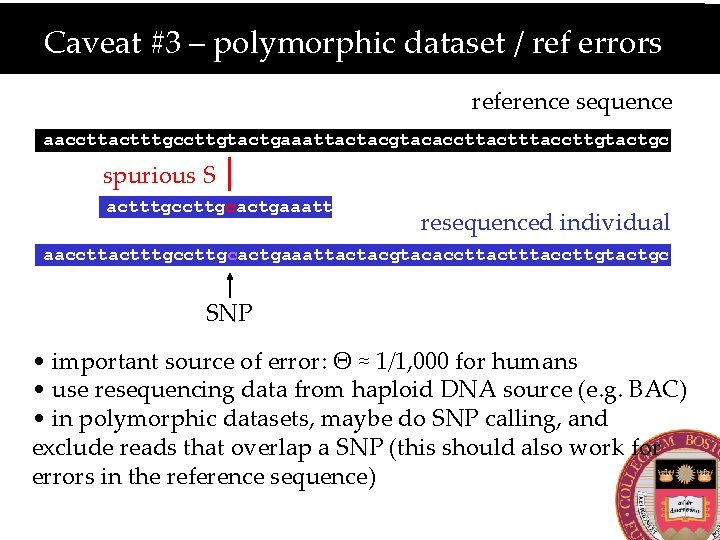
Caveat #3 – polymorphic dataset / ref errors reference sequence aaccttactttgccttgtactgaaattactacgtacaccttactttaccttgtactgc spurious S actttgccttgcactgaaatt resequenced individual aaccttactttgccttgcactgaaattactacgtacaccttactttaccttgtactgc SNP • important source of error: Θ ≈ 1/1, 000 for humans • use resequencing data from haploid DNA source (e. g. BAC) • in polymorphic datasets, maybe do SNP calling, and exclude reads that overlap a SNP (this should also work for errors in the reference sequence)

Caveat #4 – very low quality reads aaccttactttgccttgtactgaaattactacgtacaccttactttaccttgtactgc acaatgcgttgca***agatt • these reads are hard to even align elude error statistics • only tabulate error stats for reads that will be aligned?
Study design • Took 3 random lanes each from 3 random runs of PE Illumina reads from Sanger (100, 000 random pairs per lane) • Mapped the reads to NCBI build 36 using Mosaik. PE • Alignment conditions: paired-end alignments, unique-unique end-reads maximum 4 mismatches per end-read • Fraction aligned: on average, X fraction of the reads used Analysis by Derek Barnett

25000 0 80 84 88 92 96 100 104 108 112 116 120 124 128 132 136 140 144 148 152 156 160 164 168 172 176 180 184 188 192 196 200 204 208 212 216 220 224 228 232 236 240 244 248 252 256 260 264 268 272 276 280 284 288 292 296 300 Fragment length distribution Distribution of Fragment Lengths 20000 15000 10000 5000 Total Fragment Length

Base error and error type Error rate over all bases Correct 98, 76% Error 1, 24% Rates of Specific Error Types Insertions 1, 43% Substitutions 95, 34% Deletions 3, 23%

Base error rate by substitution type
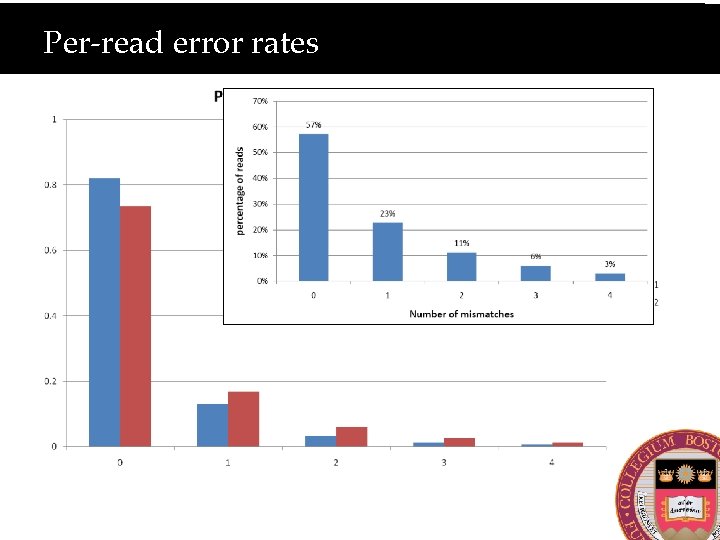
Per-read error rates
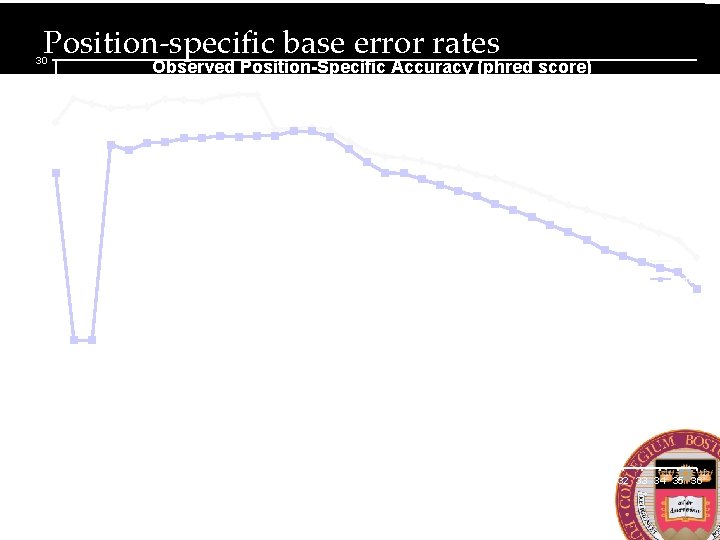
Position-specific base error rates 30 Observed Position-Specific Accuracy (phred score) 25 20 Mate 1 15 Mate 2 10 5 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36

Accuracy across lanes and runs
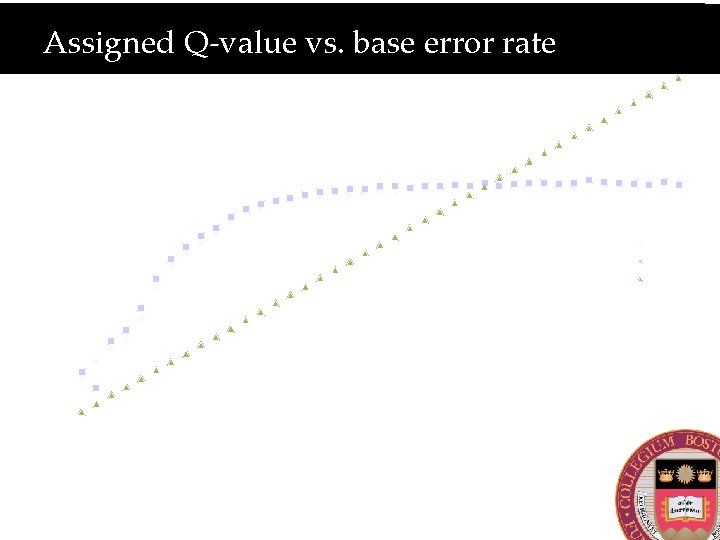
Assigned Q-value vs. base error rate 40 Observed Accuracy for Given BQ (phred score) 35 Observed Accuracy 30 25 Mate 1 20 Mate 2 Expected 15 10 5 0 0 5 10 15 20 Given BQ 25 30 35 40

Does the Q-value depend on base cycle?
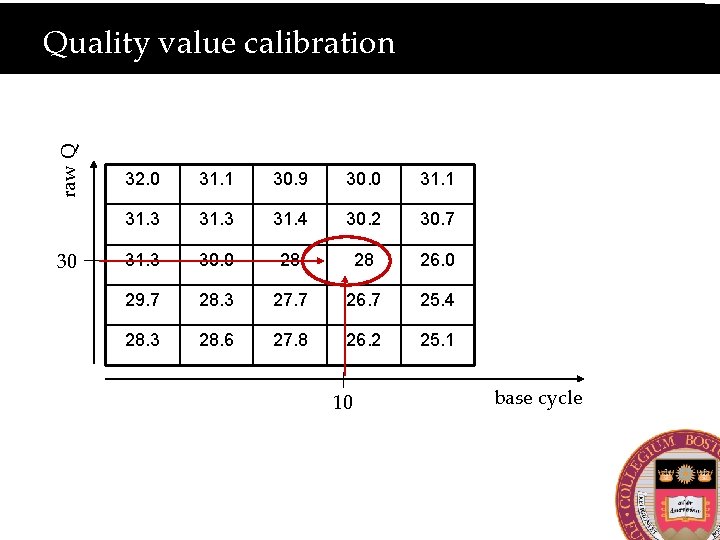
raw Q Quality value calibration 30 32. 0 31. 1 30. 9 30. 0 31. 1 31. 3 31. 4 30. 2 30. 7 31. 3 30. 0 28 28 26. 0 29. 7 28. 3 27. 7 26. 7 25. 4 28. 3 28. 6 27. 8 26. 2 25. 1 10 base cycle
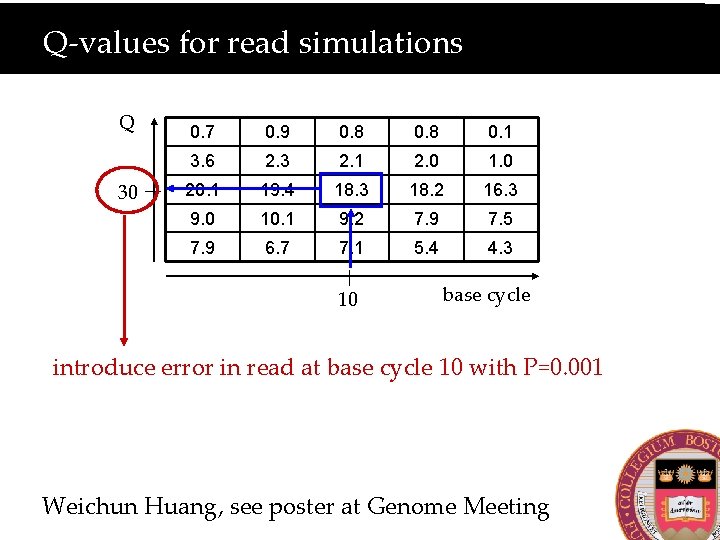
Q-values for read simulations Q 30 0. 7 0. 9 0. 8 0. 1 3. 6 2. 3 2. 1 2. 0 1. 0 20. 1 19. 4 18. 3 18. 2 16. 3 9. 0 10. 1 9. 2 7. 9 7. 5 7. 9 6. 7 7. 1 5. 4 4. 3 10 base cycle introduce error in read at base cycle 10 with P=0. 001 Weichun Huang, see poster at Genome Meeting

Thanks Michael Derek Aaron Chip Weichun
- Slides: 19