Balanced Translocation detected by FISH 1 Red Chrom
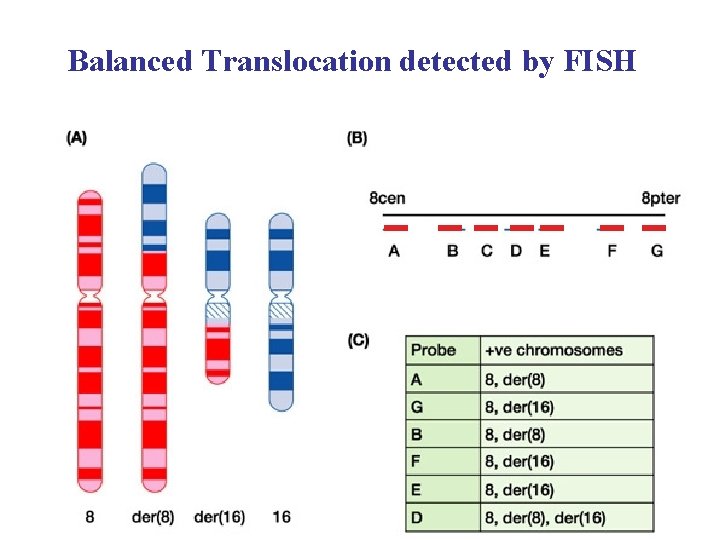
Balanced Translocation detected by FISH 1
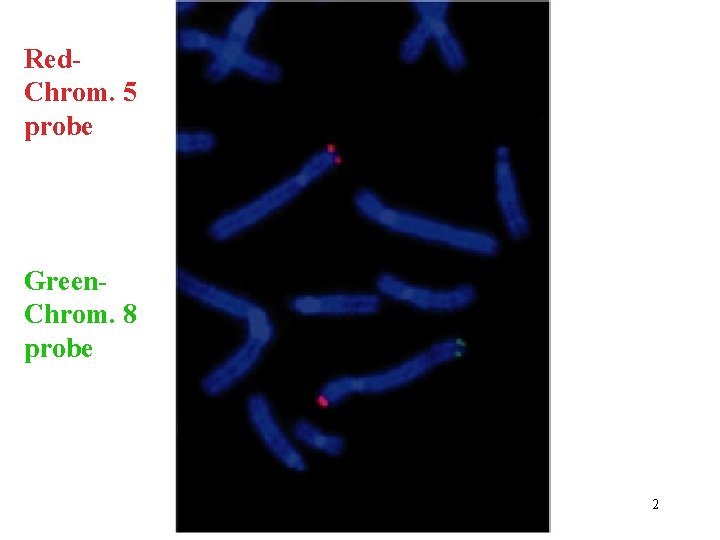
Red. Chrom. 5 probe Green. Chrom. 8 probe 2
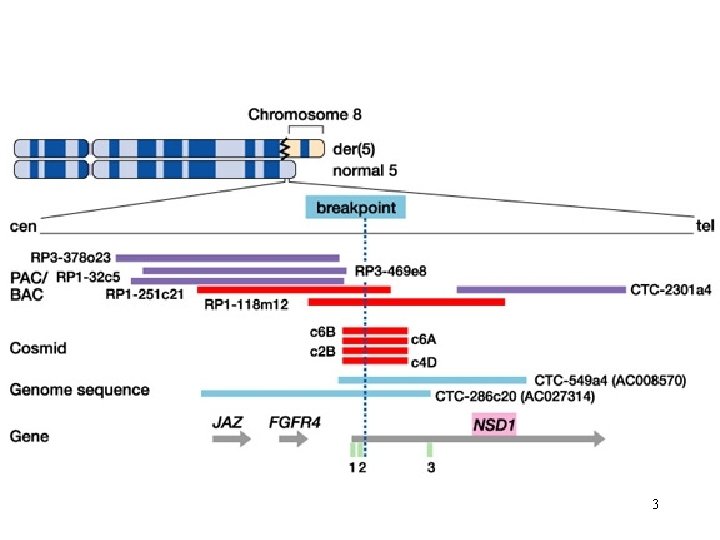
3
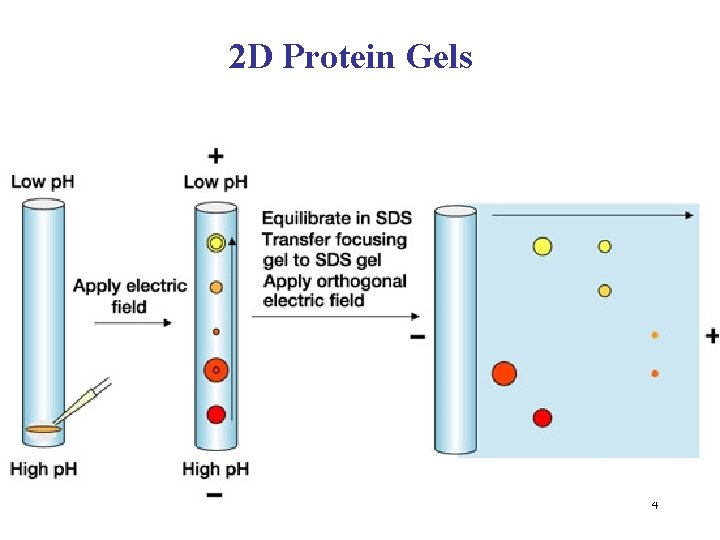
2 D Protein Gels 4

5
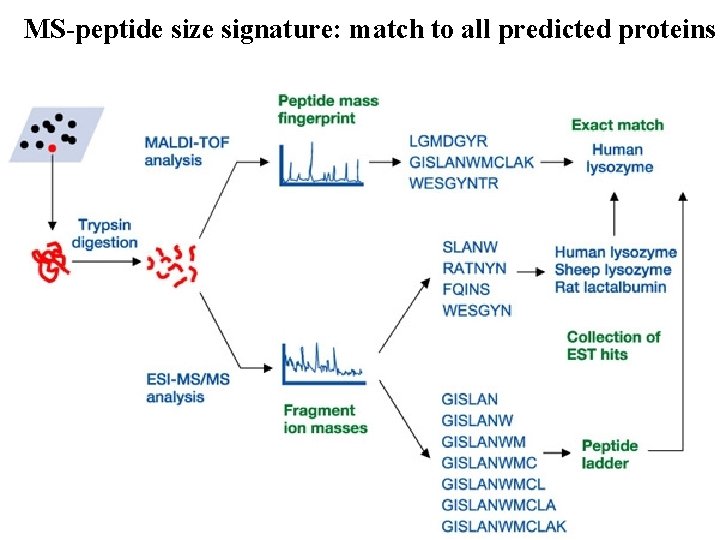
MS-peptide size signature: match to all predicted proteins 6
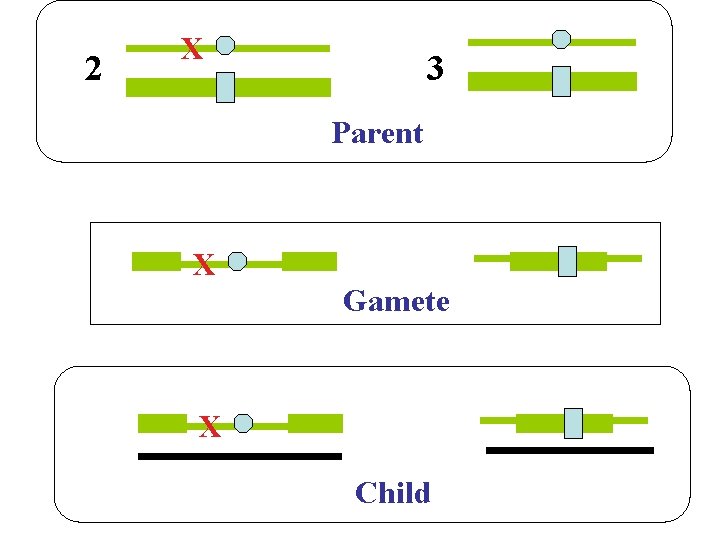
2 X 3 Parent X Gamete X Child 7
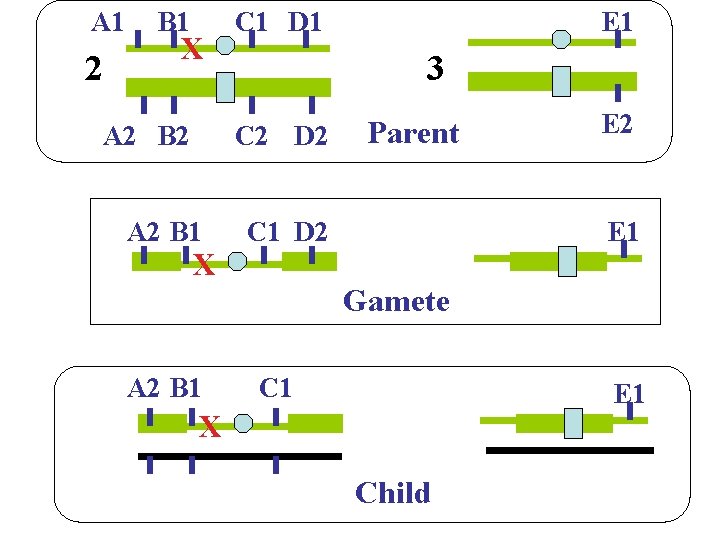
A 1 2 B 1 X A 2 B 2 C 1 D 1 3 C 2 D 2 A 2 B 1 X E 1 Parent C 1 D 2 E 1 Gamete A 2 B 1 C 1 E 1 X Child 8
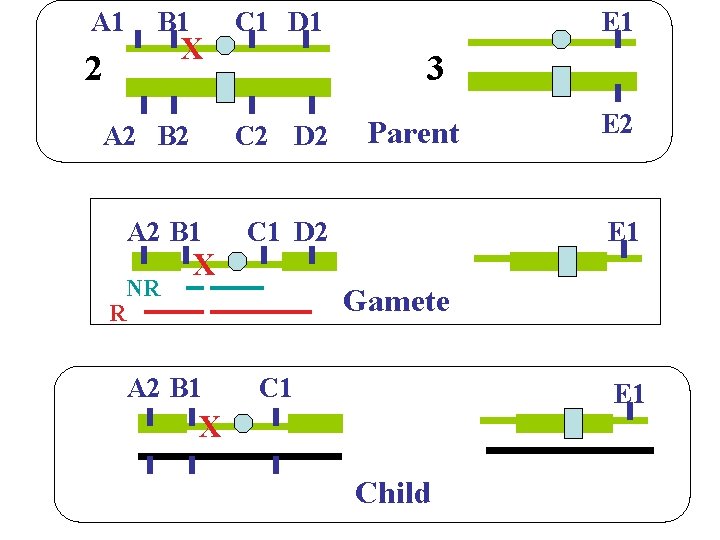
A 1 B 1 X 2 A 2 B 2 X E 1 3 C 2 D 2 A 2 B 1 NR R C 1 D 1 Parent C 1 D 2 E 1 Gamete A 2 B 1 C 1 E 1 X Child 9

Positional Cloning by Recombination Mapping 1. Follow the mutation 2. Follow which DNAs are co-inherited (linked) 10
Positional Cloning by Recombination Mapping 1. Follow the mutation To determine disease gene presence or absence (genotype) from phenotype you must first establish Dominant / recessive Aurosomal / sex-linked 11
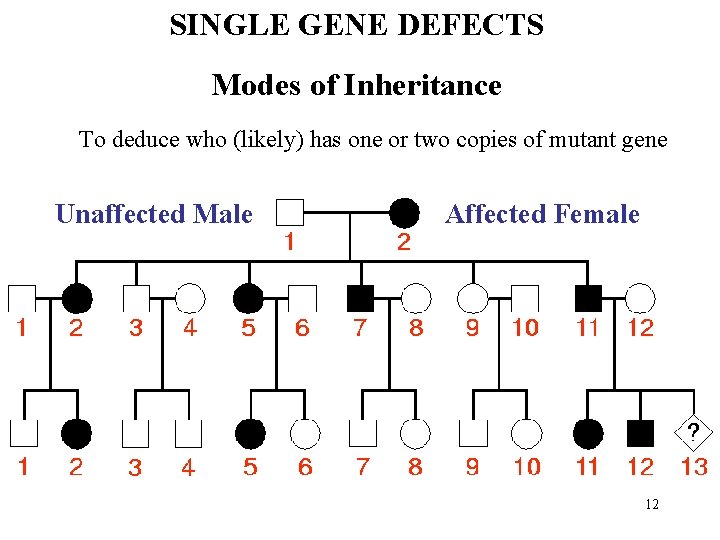
SINGLE GENE DEFECTS Modes of Inheritance To deduce who (likely) has one or two copies of mutant gene Unaffected Male Affected Female 12
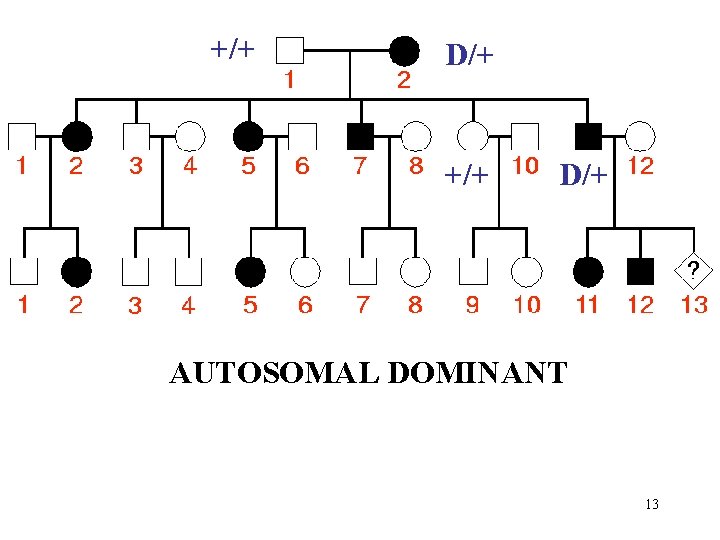
+/+ D/+ AUTOSOMAL DOMINANT 13
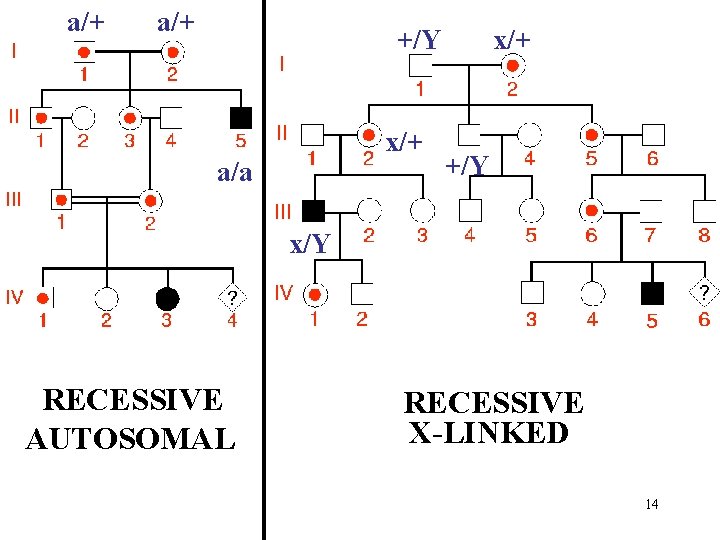
a/+ +/Y x/+ a/a x/+ +/Y x/Y RECESSIVE AUTOSOMAL RECESSIVE X-LINKED 14
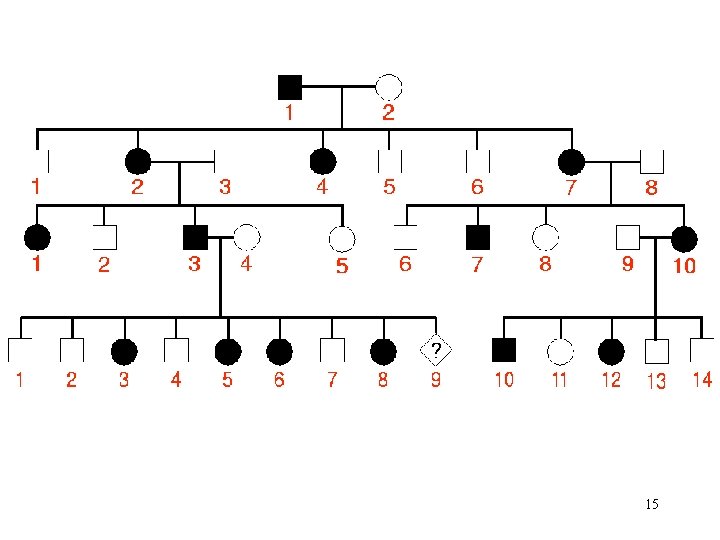
15
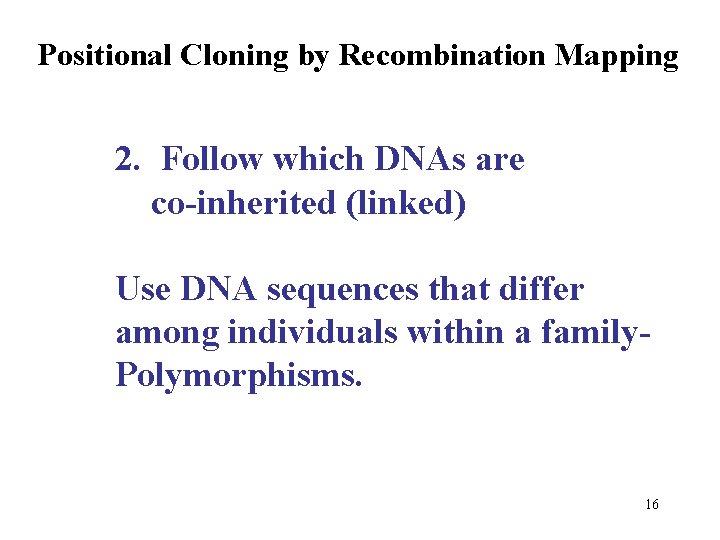
Positional Cloning by Recombination Mapping 2. Follow which DNAs are co-inherited (linked) Use DNA sequences that differ among individuals within a family. Polymorphisms. 16
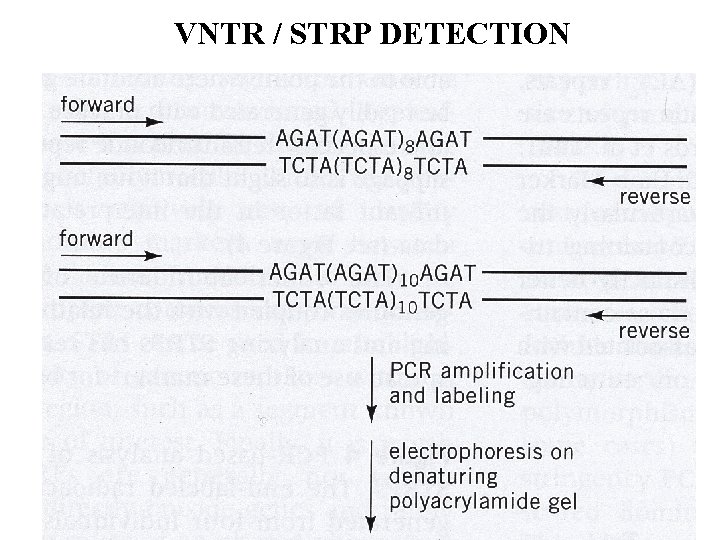
VNTR / STRP DETECTION 17
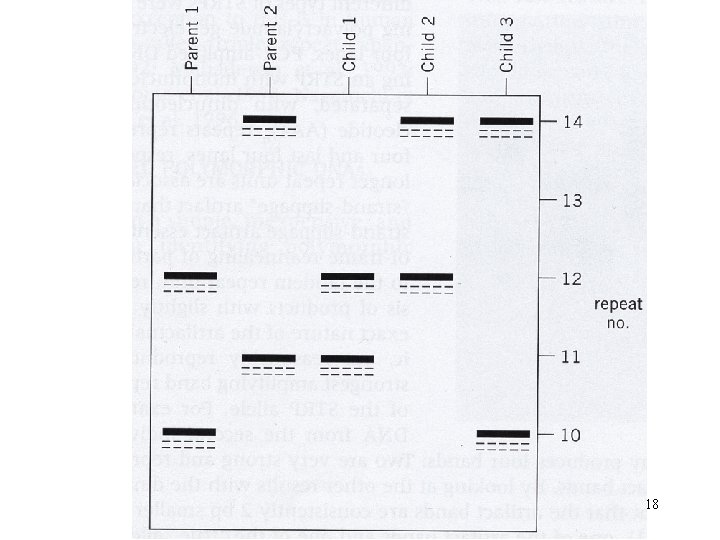
18
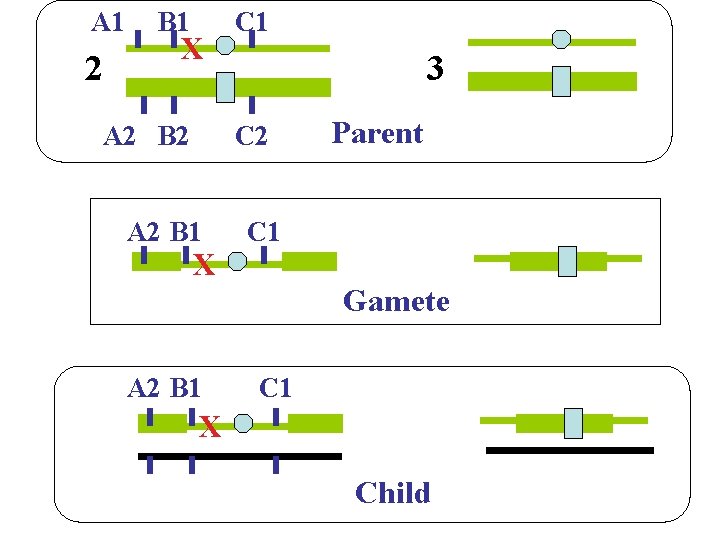
A 1 2 B 1 X A 2 B 2 C 1 3 C 2 A 2 B 1 X Parent C 1 Gamete A 2 B 1 C 1 X Child 19
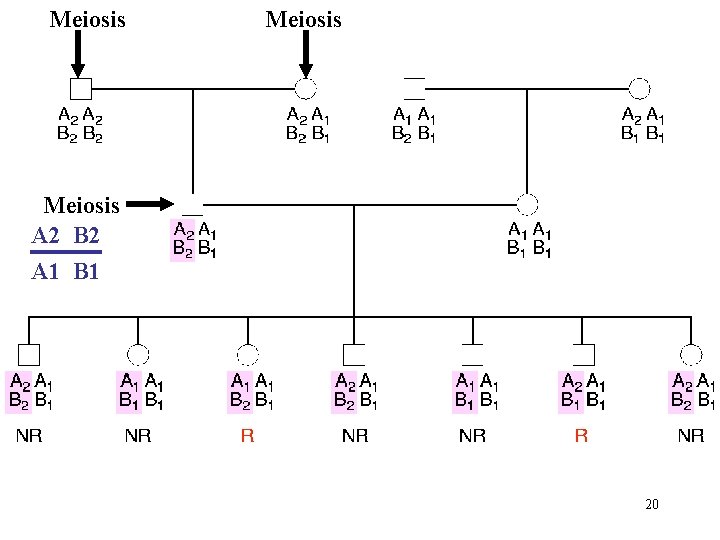
Meiosis A 2 B 2 A 1 B 1 20
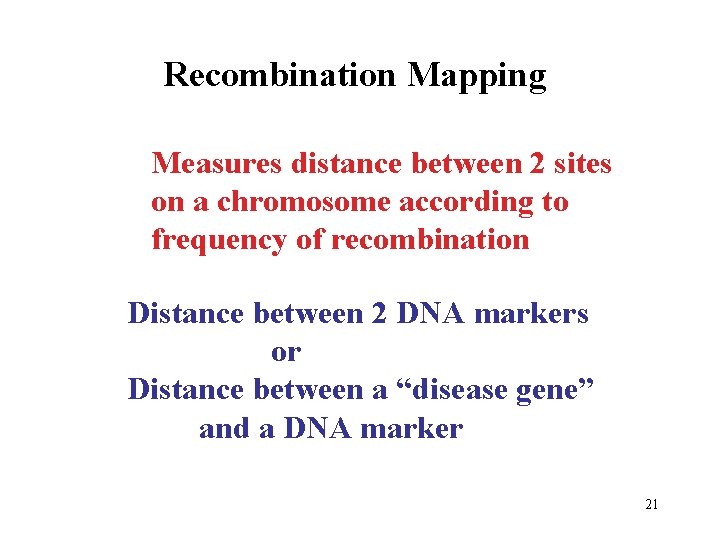
Recombination Mapping Measures distance between 2 sites on a chromosome according to frequency of recombination Distance between 2 DNA markers or Distance between a “disease gene” and a DNA marker 21
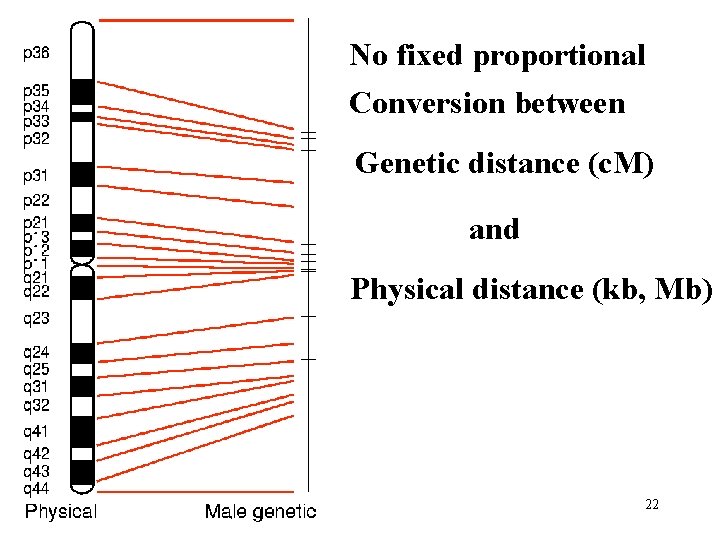
No fixed proportional Conversion between Genetic distance (c. M) and Physical distance (kb, Mb) 22
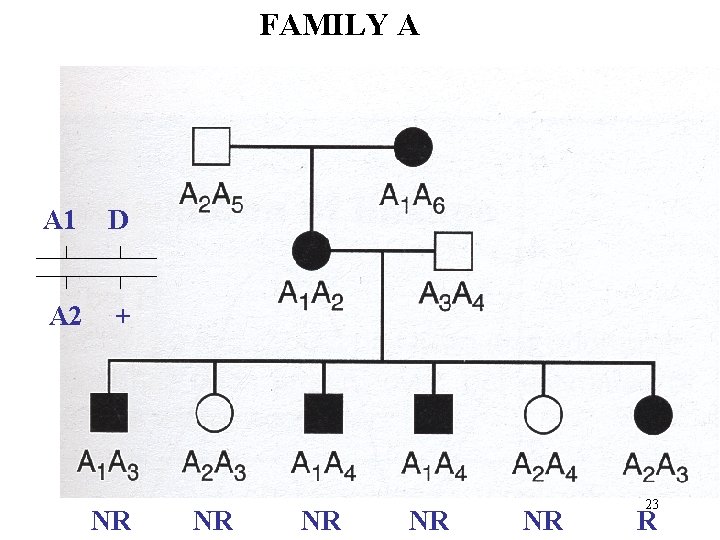
FAMILY A A 1 D A 2 + NR NR NR 23 R
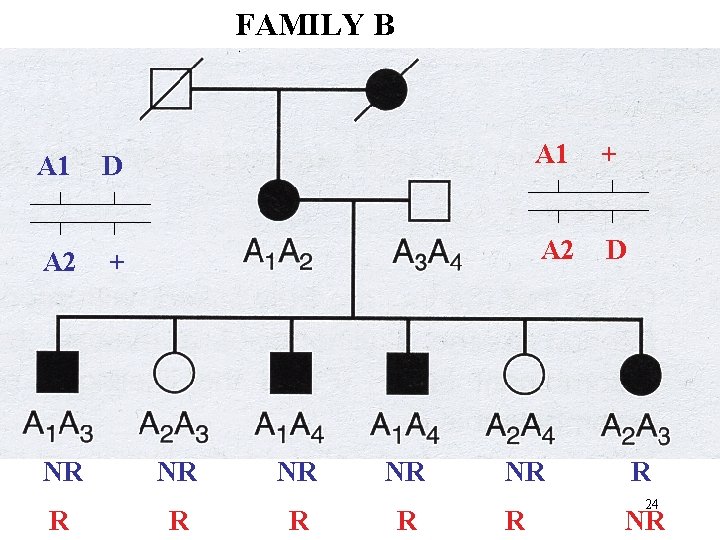
FAMILY B A 1 D A 1 + A 2 D NR R NR R R 24 NR

INFORMATIVE MEIOSIS Ideally: unambiguous inheritance of mutation and markers (requires heterozygosity for each in parent) knowledge of which alleles linked in parent (phase) 25

Assign numbers to results of linkage analysis to deal with non-ideal meioses to sum data from many meioses in a family to sum data from several families 26
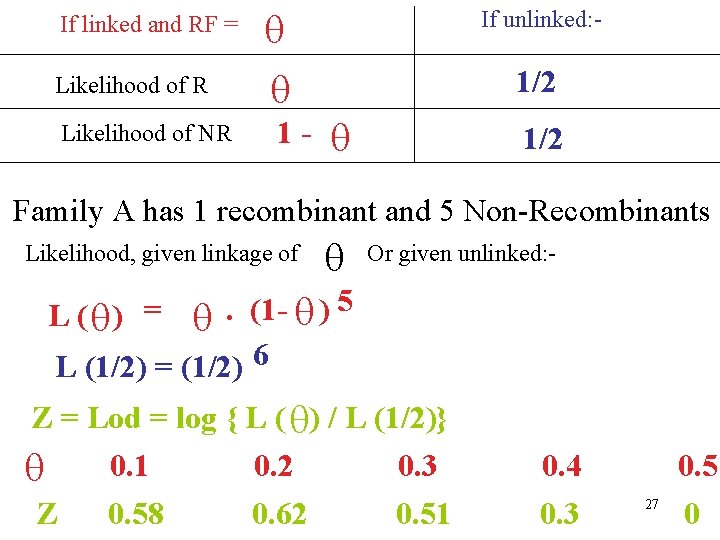
If linked and RF = Likelihood of R Likelihood of NR 1 - If unlinked: - 1/2 Family A has 1 recombinant and 5 Non-Recombinants Likelihood, given linkage of Or given unlinked: - 5. (1) L (1/2) = (1/2) 6 L ( ) = Z = Lod = log { L ( ) / L (1/2)} 0. 1 0. 2 0. 3 Z 0. 58 0. 62 0. 51 0. 4 0. 3 27 0. 5 0
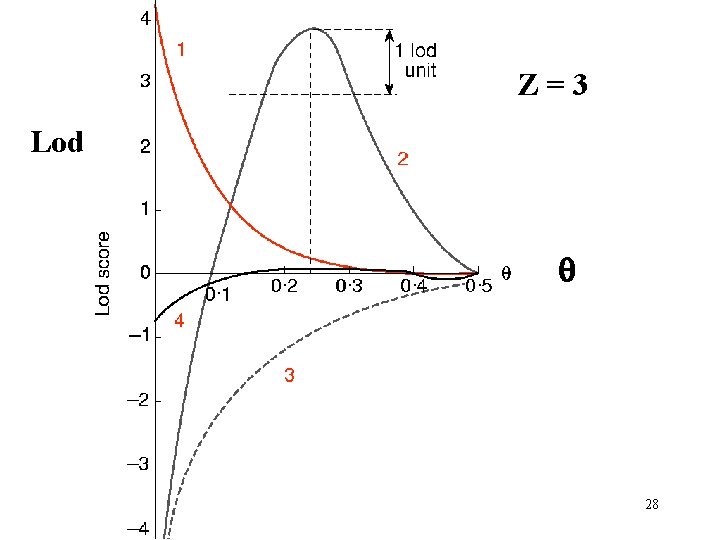
Z=3 Lod q 28
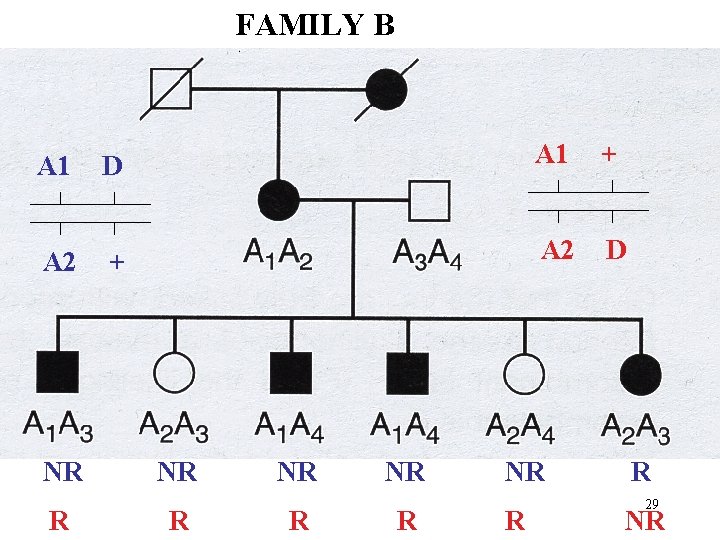
FAMILY B A 1 D A 1 + A 2 D NR R NR R R 29 NR
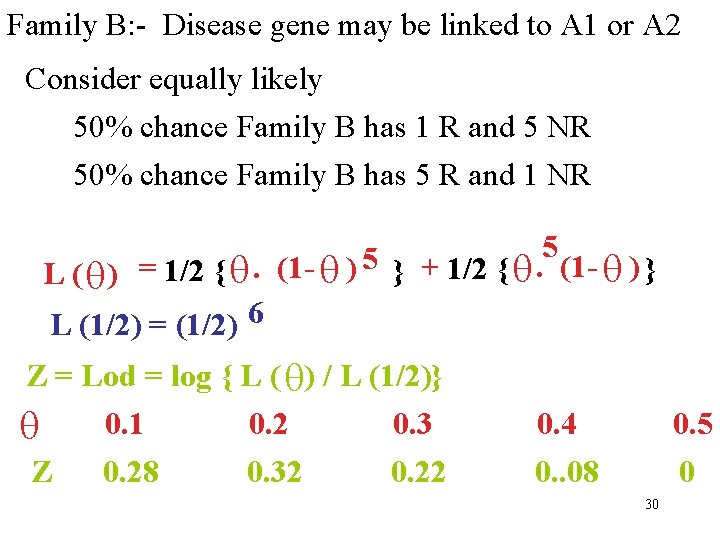
Family B: - Disease gene may be linked to A 1 or A 2 Consider equally likely 50% chance Family B has 1 R and 5 NR 50% chance Family B has 5 R and 1 NR 5 5 L ( ) = 1/2 { . (1 - ) } + 1/2 { . (1 - ) } L (1/2) = (1/2) 6 Z = Lod = log { L ( ) / L (1/2)} 0. 1 0. 2 0. 3 Z 0. 28 0. 32 0. 22 0. 4 0. . 08 0. 5 0 30
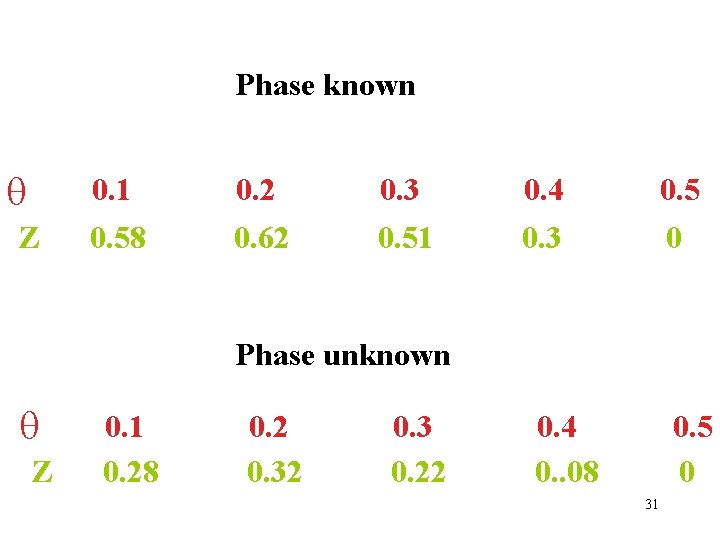
Phase known Z 0. 1 0. 58 0. 2 0. 62 0. 3 0. 51 0. 4 0. 3 0. 5 0 Phase unknown Z 0. 1 0. 28 0. 2 0. 3 0. 22 0. 4 0. . 08 0. 5 0 31

For family “A” with meioses 1, 2, 3, 4 …. . Z = Z 1 + Z 2 + Z 3 + Z 4 +…. . For multiple families, “A”, “B”, “C”, “D”…. . Z = Z(A) + Z(B) + Z(C) + Z(D) + …. Assumption: same gene responsible for disease in all families Problem: locus heterogeneity 32
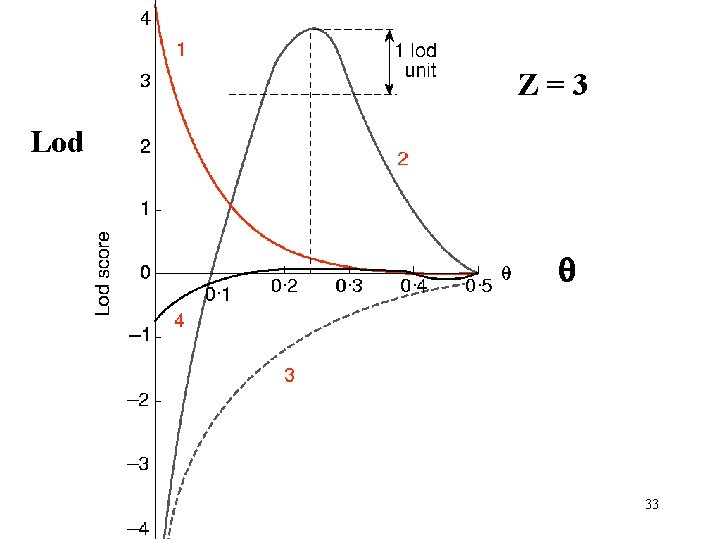
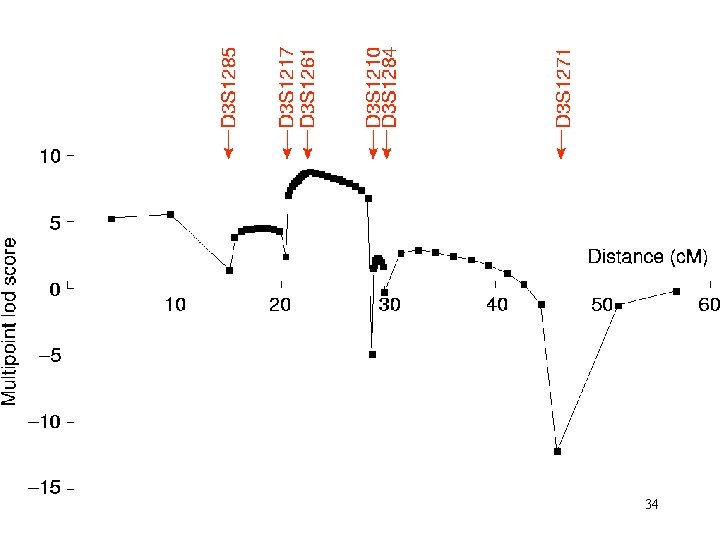
Z=3 Lod q 33
34
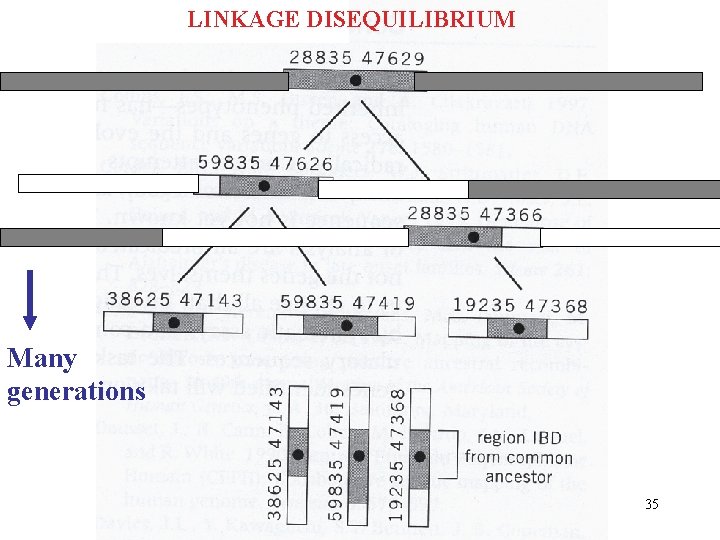
LINKAGE DISEQUILIBRIUM Many generations 35
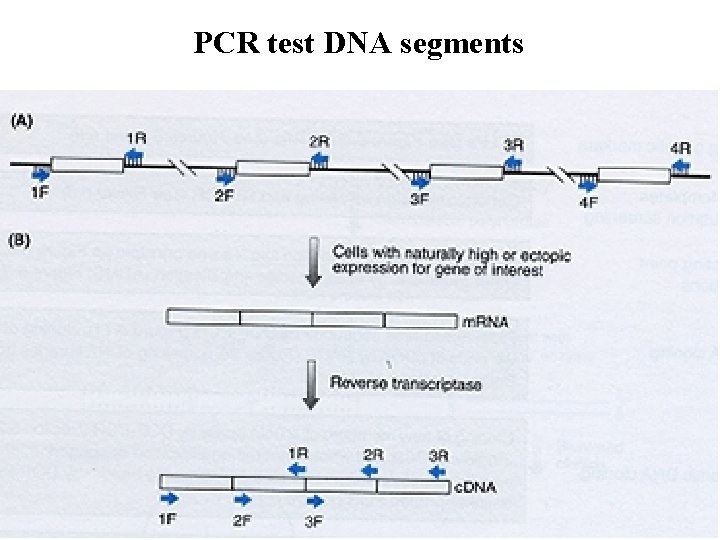
PCR test DNA segments 36
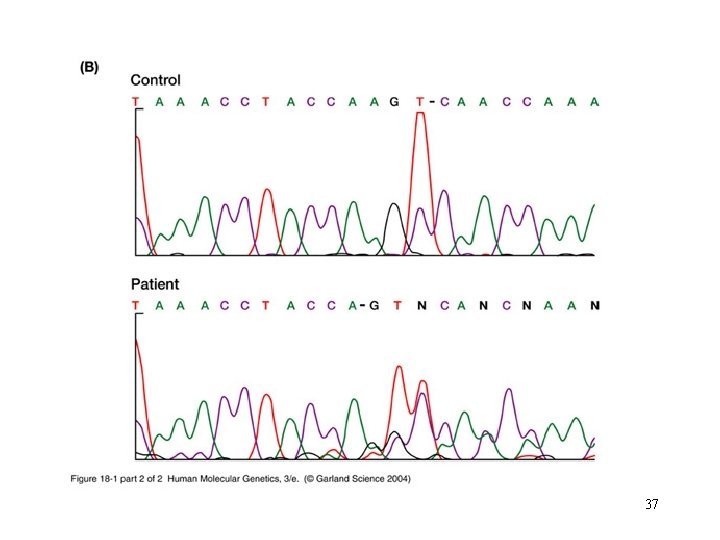
37

Testing for specific mutations 38
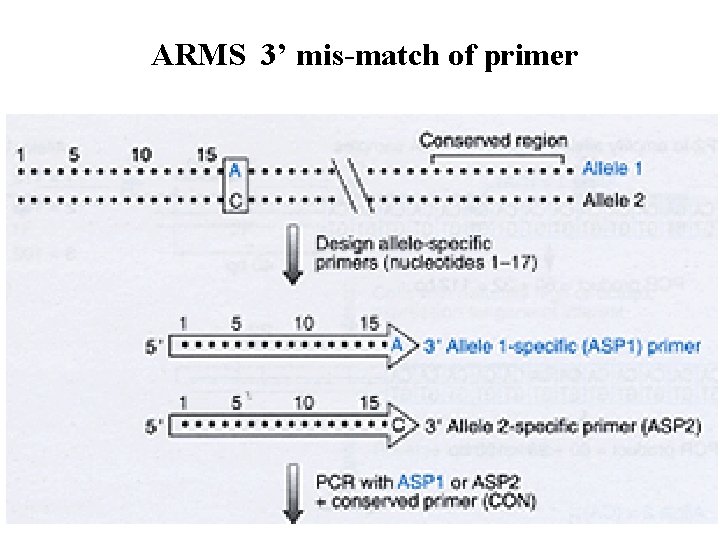
ARMS 3’ mis-match of primer 39
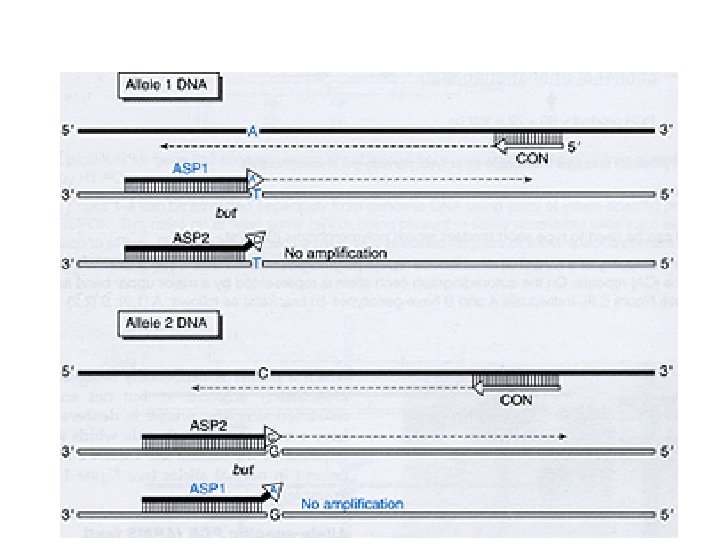
40
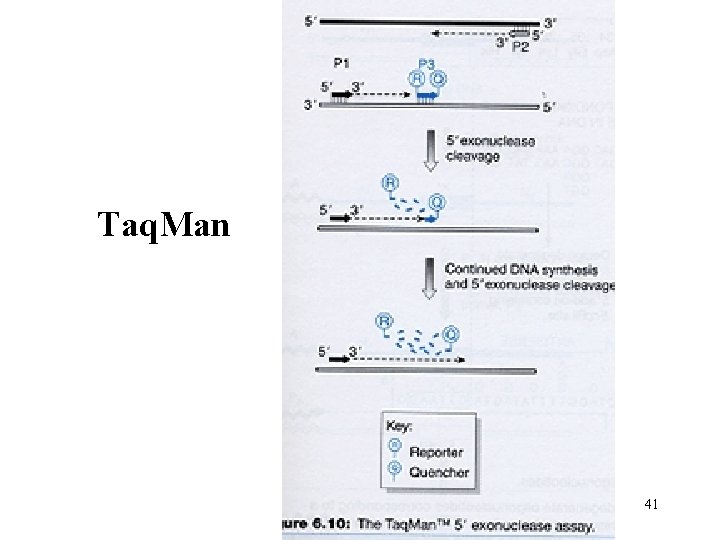
Taq. Man 41
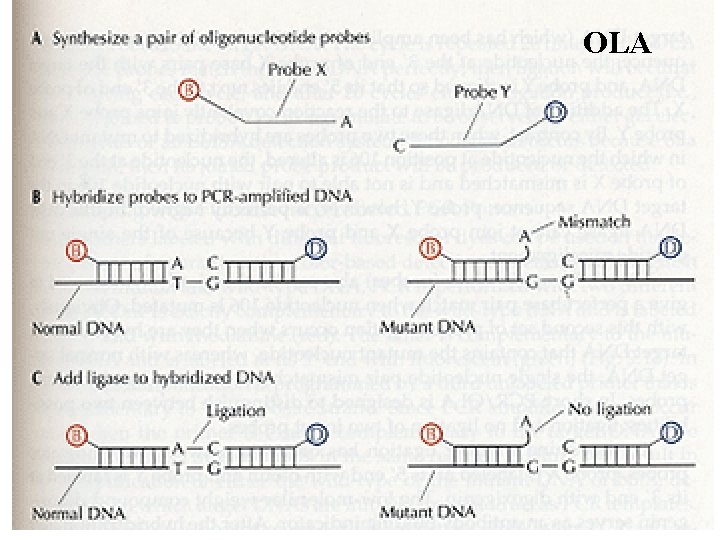
OLA 42
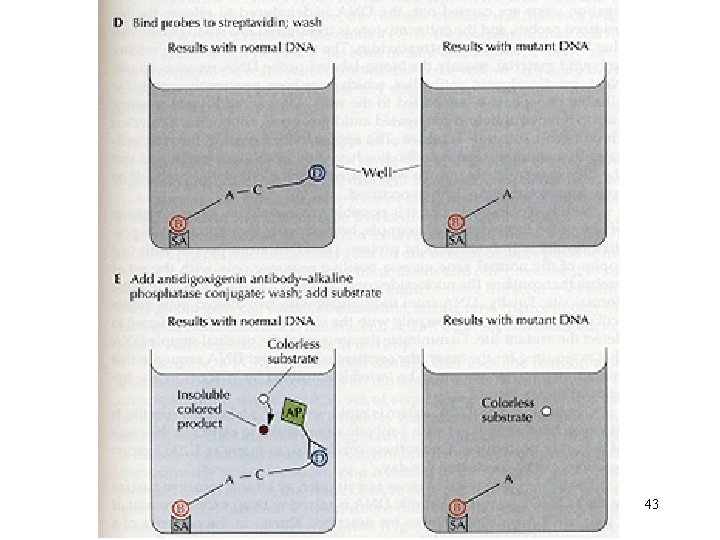
43
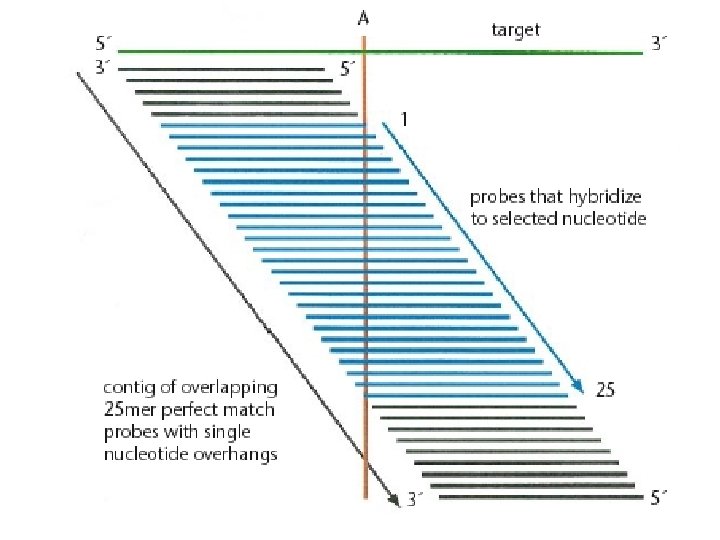
44
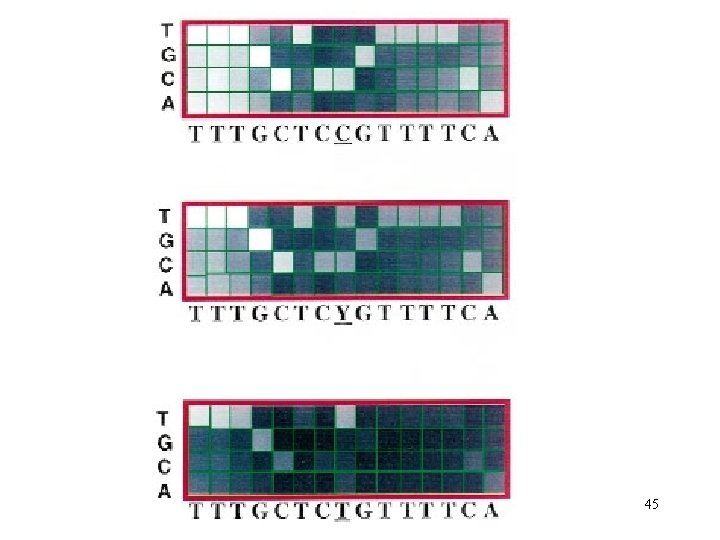
45
- Slides: 45