BACTERIOLOGY 3 BACTERIAL CELL STRUCTURE b Prof Dr

BACTERIOLOGY (3) BACTERIAL CELL STRUCTURE (b) Prof. Dr. Waiel Farghaly Professor of Microbiology/ Dept. Botany, Fac. Science
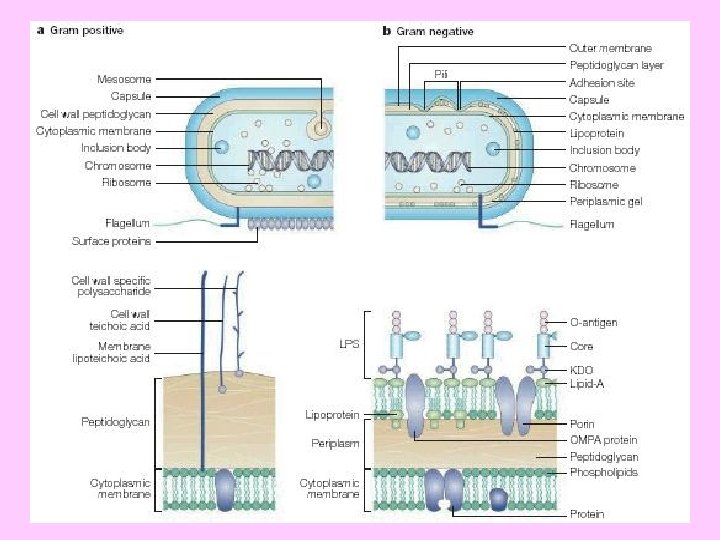
THE CELL ENVELOPES • The “cell envelopes” are the several layers of material that envelope or enclose the protoplasm of the cell. The cell protoplasm (cytoplasm + membrane) is surrounded by the plasma membrane, a cell wall and a capsule. • Almost all prokaryotes have a cell wall to prevent damage to the underlying protoplast. • The wall is rigid but ductile. 1 - It gives the cell the required mechanical strength 2 - Determines the cell shape and encloses the cytoplasm 3 - The characteristic antigens of each bacterium are also located on their cell walls.
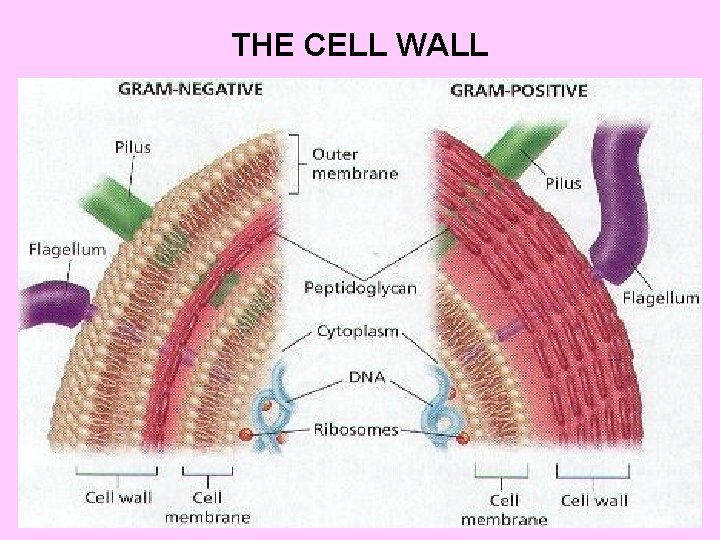
THE CELL WALL

The presence of the cell wall can be evidenced by many ways: • • • Staining with special stains: mordant a heat- fixed film with tannic acid (5 - 10%), washing with distilled water (protein is altered and will not take up the stain), the wall can be stained then with 0. 2% aqueous crystal violet. Plasmolysis of the cell: cell contents will contract in hypertonic solution leaving the wall without contraction and then, can be stained easily. Cell destruction by various methods: by ultrasonic waves in the presence of powdered glass, or autolysis and digestion of the protoplasm without affecting the wall. Then wall can be separated from the destroyed cell by centrifugation. Investigation of wall preparations can be carried out by electron microscope.
Cell walls are unique structures 1. They are essential structures for viability, as described above. 2. They are composed of unique components found nowhere else in nature. 3. They are one of the most important sites for attack by antibiotics. 4. They provide ligands for adherence and receptor sites for drugs or viruses. 5. They cause symptoms of disease in animals. 6. They provide the immunological distinction and variation among strains of bacteria.
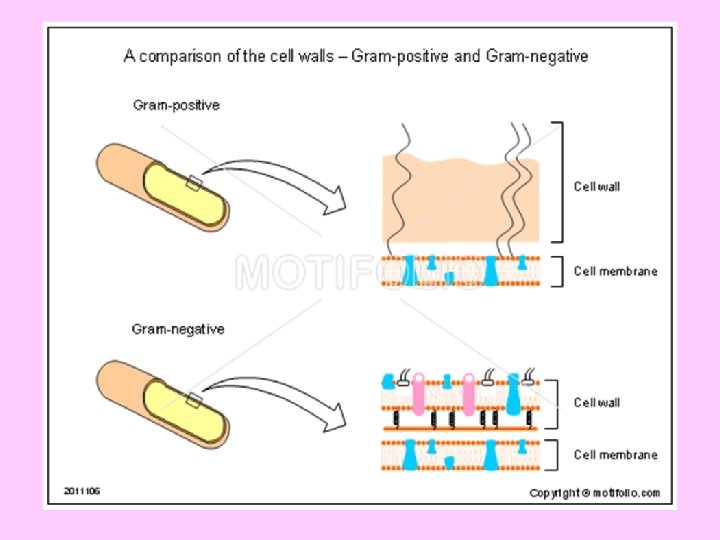
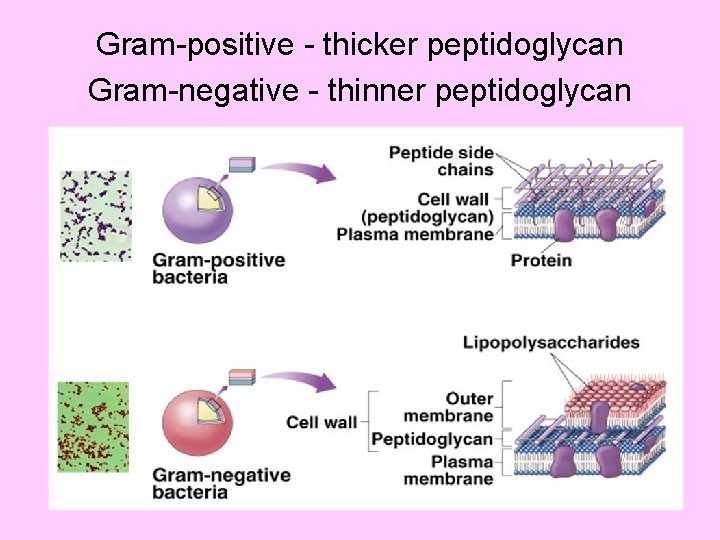
Gram-positive - thicker peptidoglycan Gram-negative - thinner peptidoglycan
Peptidoglycan (murein, mucopeptide, mucocomplex. . etc) • Peptidoglycan is a polymer of disaccharides (a glycan) cross-linked by short chains of amino acids (peptides), and many types of peptidoglycan exist. • All bacterial peptidoglycans contain N-acetylmuramic acid, which is the definitive component of murein. • The cell walls of Archaea may be composed of protein, polysaccharides, or peptidoglycan- like molecules, but never contain murein. This feature distinguishes the Bacteria from the Archaea.
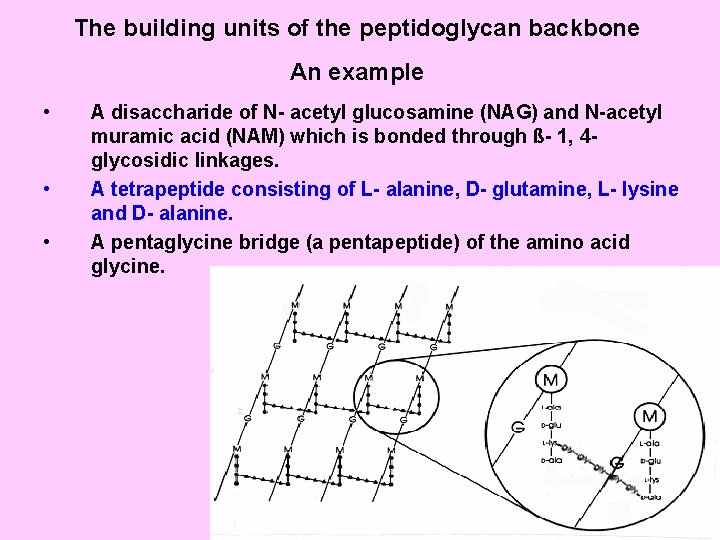
The building units of the peptidoglycan backbone An example • • • A disaccharide of N- acetyl glucosamine (NAG) and N-acetyl muramic acid (NAM) which is bonded through ß- 1, 4 glycosidic linkages. A tetrapeptide consisting of L- alanine, D- glutamine, L- lysine and D- alanine. A pentaglycine bridge (a pentapeptide) of the amino acid glycine.
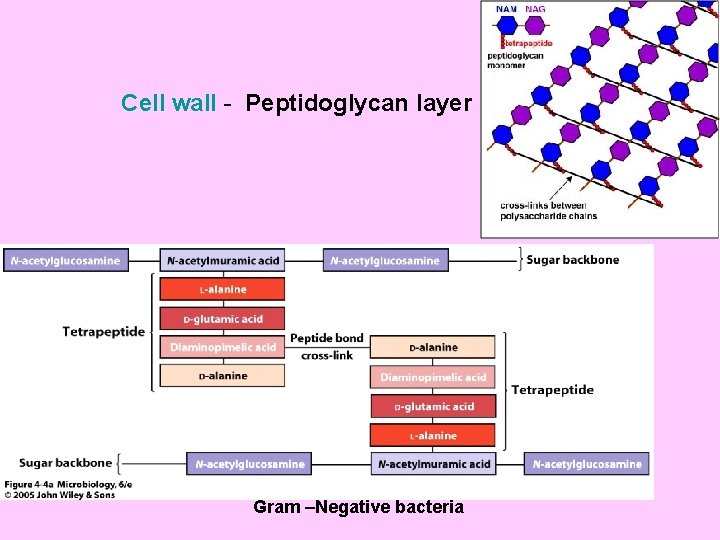
Cell wall - Peptidoglycan layer Gram –Negative bacteria
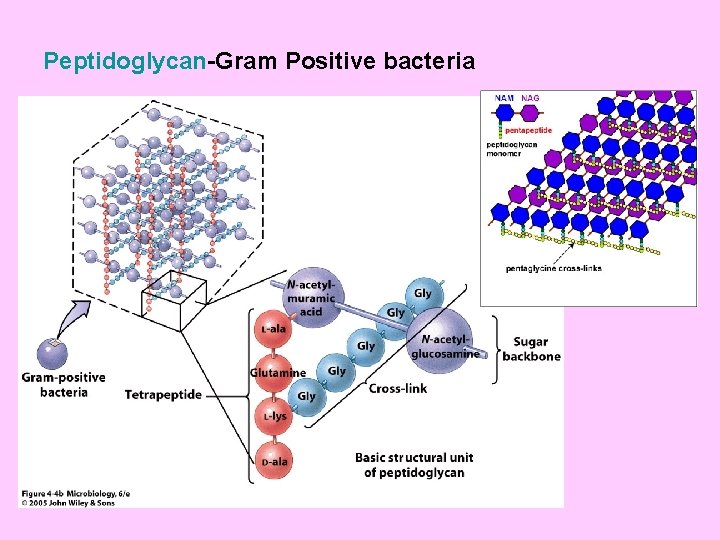
Peptidoglycan-Gram Positive bacteria
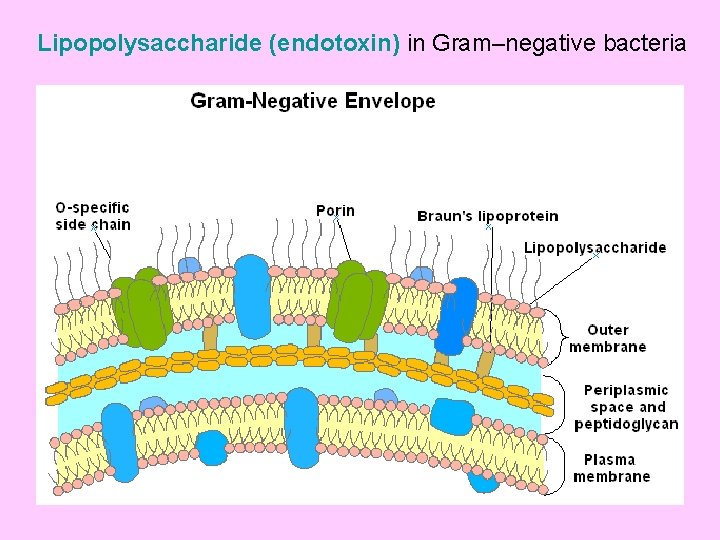
Lipopolysaccharide (endotoxin) in Gram–negative bacteria
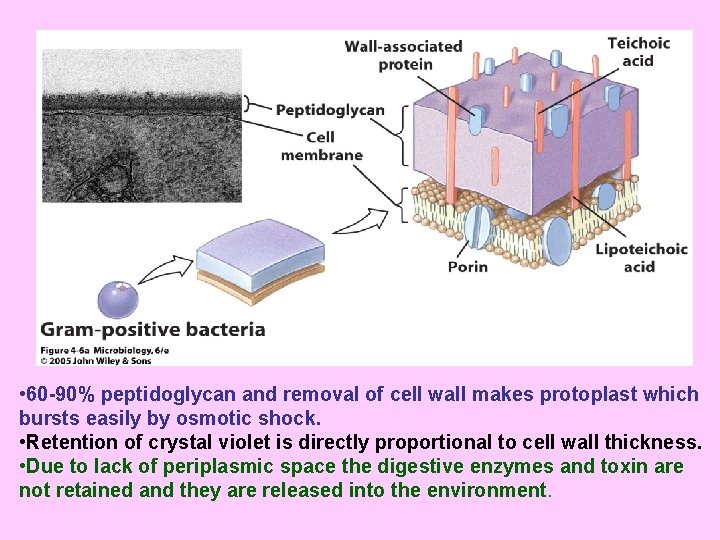
• 60 -90% peptidoglycan and removal of cell wall makes protoplast which bursts easily by osmotic shock. • Retention of crystal violet is directly proportional to cell wall thickness. • Due to lack of periplasmic space the digestive enzymes and toxin are not retained and they are released into the environment.
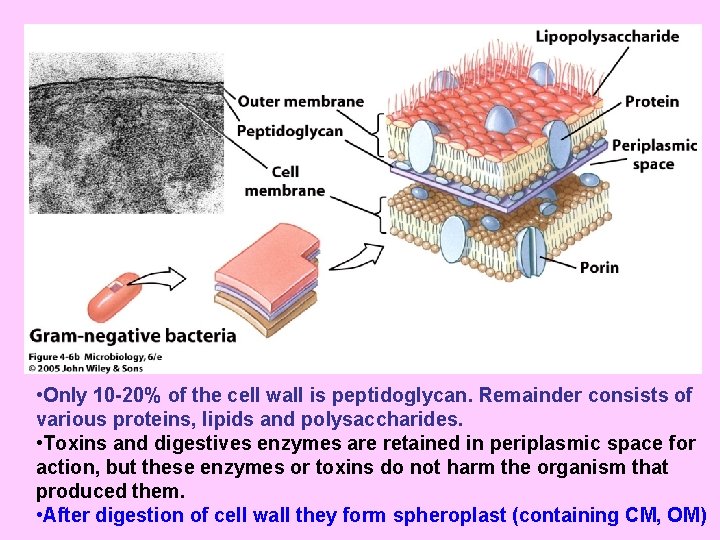
• Only 10 -20% of the cell wall is peptidoglycan. Remainder consists of various proteins, lipids and polysaccharides. • Toxins and digestives enzymes are retained in periplasmic space for action, but these enzymes or toxins do not harm the organism that produced them. • After digestion of cell wall they form spheroplast (containing CM, OM)

Mycobacteria • Cell wall is thick, contains 60% lipids and very less peptidoglycan. • The lipids make acid- fast organisms impermeable to most other strains and protect them from acids and alkali’s. • The organism grows slowly because the lipids impede the entry of nutrients into cell and cells spend large amount of energy to synthesize lipids. • They can be stained by Gram staining method and stained as Gram positive.
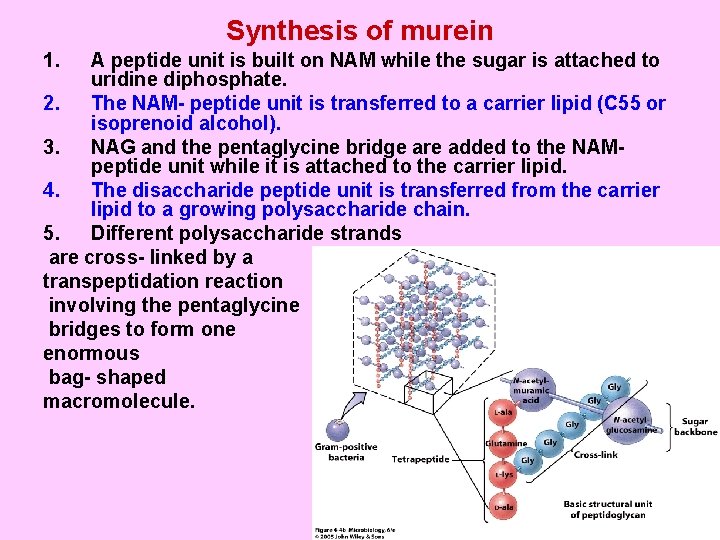
Synthesis of murein 1. A peptide unit is built on NAM while the sugar is attached to uridine diphosphate. 2. The NAM- peptide unit is transferred to a carrier lipid (C 55 or isoprenoid alcohol). 3. NAG and the pentaglycine bridge are added to the NAMpeptide unit while it is attached to the carrier lipid. 4. The disaccharide peptide unit is transferred from the carrier lipid to a growing polysaccharide chain. 5. Different polysaccharide strands are cross- linked by a transpeptidation reaction involving the pentaglycine bridges to form one enormous bag- shaped macromolecule.
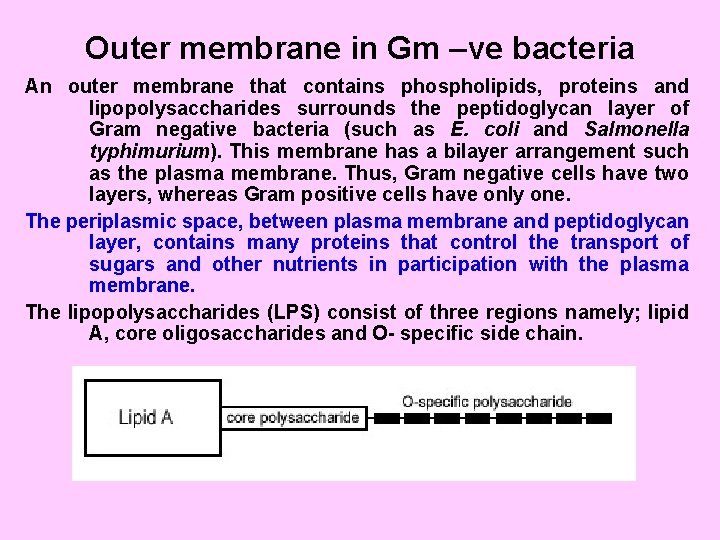
Outer membrane in Gm –ve bacteria An outer membrane that contains phospholipids, proteins and lipopolysaccharides surrounds the peptidoglycan layer of Gram negative bacteria (such as E. coli and Salmonella typhimurium). This membrane has a bilayer arrangement such as the plasma membrane. Thus, Gram negative cells have two layers, whereas Gram positive cells have only one. The periplasmic space, between plasma membrane and peptidoglycan layer, contains many proteins that control the transport of sugars and other nutrients in participation with the plasma membrane. The lipopolysaccharides (LPS) consist of three regions namely; lipid A, core oligosaccharides and O- specific side chain.
• The lipid A moiety is hydrophobic, while the core oligosaccharides and O- side chain are highly hydrophilic • The oligosaccharide region is composed of ten sugar units that project outwards followed by the O- side chain (repeating tetrasaccharide units). • The two sugar regions contain several sugars rarely present elsewhere in nature (e. g. 2 - keto- 3 - deoxyoctonate (KDO), an eight carbon sugar; heptose, a seven- carbon sugar and abequose). • The LPS molecule is negatively charged because several of its sugars are phosphorylated • The sequence of OM synthesis starts with lipid A, core oligosaccharides and, finally, O- side chain is added to the tip of the molecule. • The core oligosaccharide is synthesized by sequential addition of sugars from activated donors such as UDP- glucose and UDPgalactose. While the O- side chain synthesis is similar to murein. Its repeating tetrasaccharide units are built on the inner leaflet of the plasma membrane and transferred to the growing chain by the same carrier lipid (C 55).
Gram –ve bacteria counteracts host defenses 1. The saturated fatty acid chains (lipid A) contribute to the barrier role of the outer membrane (periplasmic proteins kept in and harmful molecules kept out) - penicillin does not enter Gram –ve cells. 2. The lipid A may also confer rigidity on the outer membrane, while Oside chains are not essential for viability (some E. coli do not have it). 3. The outer coat of polysaccharide makes the bacterial surface very hydrophilic which reduces susceptibility to phagocytosis by host cells. 4. The O- specific chains of LPS are highly diverse. Gram negative bacteria can mutate rapidly to alter the nature of these chains. A host population not exposed to the new surface structure will have a low level of antibody against it. Thus, varying the O- side chains is a way of staying one step ahead of the host’s defense. 5. The genetic information for the alteration of O- side chains sometimes comes from a temperate phage hidden by the Gram negative bacterium (e. g. phage P 22 contributes a gene that adds glucose to the repeated tetrasaccharide unit). This kind of alteration is called phage conversion.
- Slides: 20