Avogadro Scale Engineering COMPLEXITY Physics of Information Technology
![Homework • I] Nanotech Design: Find an error function for which it is optimal Homework • I] Nanotech Design: Find an error function for which it is optimal](https://slidetodoc.com/presentation_image_h2/61179f5eed7e860bea77885f2f547fe3/image-2.jpg)
![Designing Life I] Fault Tolerant Redundancy Gene 1 Gene 2 Gene 3 Designing Life I] Fault Tolerant Redundancy Gene 1 Gene 2 Gene 3](https://slidetodoc.com/presentation_image_h2/61179f5eed7e860bea77885f2f547fe3/image-5.jpg)
![Approach 1 b] Redundant Genomes Deinococcus radiodurans (3. 2 Mb, 4 -10 Copies of Approach 1 b] Redundant Genomes Deinococcus radiodurans (3. 2 Mb, 4 -10 Copies of](https://slidetodoc.com/presentation_image_h2/61179f5eed7e860bea77885f2f547fe3/image-8.jpg)
![II] Coding m. RNA Ribosome Amino Acid II] Coding m. RNA Ribosome Amino Acid](https://slidetodoc.com/presentation_image_h2/61179f5eed7e860bea77885f2f547fe3/image-11.jpg)
![Physics of Information Technology MIT – Spring 2006 4/10 1] Von Neumann / Mc. Physics of Information Technology MIT – Spring 2006 4/10 1] Von Neumann / Mc.](https://slidetodoc.com/presentation_image_h2/61179f5eed7e860bea77885f2f547fe3/image-14.jpg)



![HOMEWORK – DUE 5/1/06 1] Consider biological cells which are able to copy their HOMEWORK – DUE 5/1/06 1] Consider biological cells which are able to copy their](https://slidetodoc.com/presentation_image_h2/61179f5eed7e860bea77885f2f547fe3/image-30.jpg)
- Slides: 30

Avogadro Scale Engineering ‘COMPLEXITY’ Physics of Information Technology MIT – Spring 2006 PART II jacobson@media. mit. edu
![Homework I Nanotech Design Find an error function for which it is optimal Homework • I] Nanotech Design: Find an error function for which it is optimal](https://slidetodoc.com/presentation_image_h2/61179f5eed7e860bea77885f2f547fe3/image-2.jpg)
Homework • I] Nanotech Design: Find an error function for which it is optimal to divide a logic area A into more than one redundant sub-Areas. • II] Design Life: (a) Design a biological system which self replicates with error correction (either genome copy redundancy with majority voting or error correcting coding). Assume the copying of each nucleotide is consumptive of one unit of energy. Show the tradeoff between energy consumption and copy fidelity. (b) Comment on the choice biology has taken (64 -3 nucleotide) codons coding for 20 amino acids. Why has biology chosen this encoding? What metric does it optimize? Could one build a biological system with 256 – 4 bit codons? Questions: jacobson@media. mit. edu

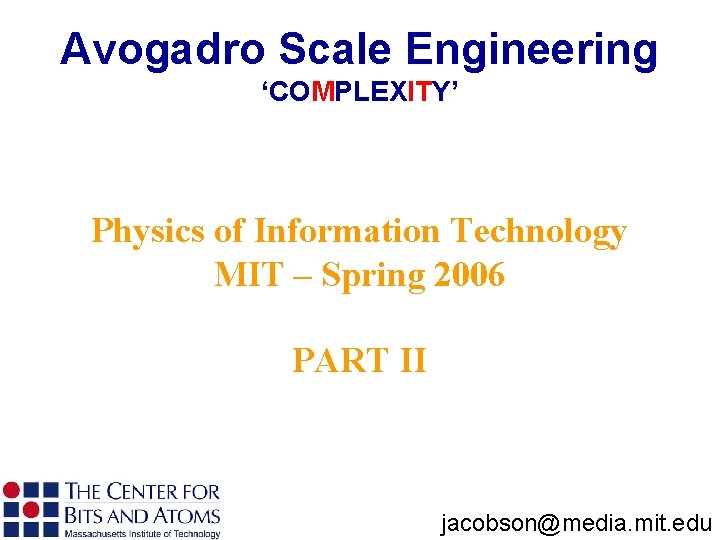
Scaling Properties of Redundant Logic (to first order) P A Probability of correct functionality = p[A] ~ e A (small A) Area = A P 1 = p[A] = e A P 2 = 2 p[A/2](1 -p[A/2])+p[A/2]2 Area = 2*A/2 = e. A –(e. A)2/4 Conclusion: P 1 > P 2

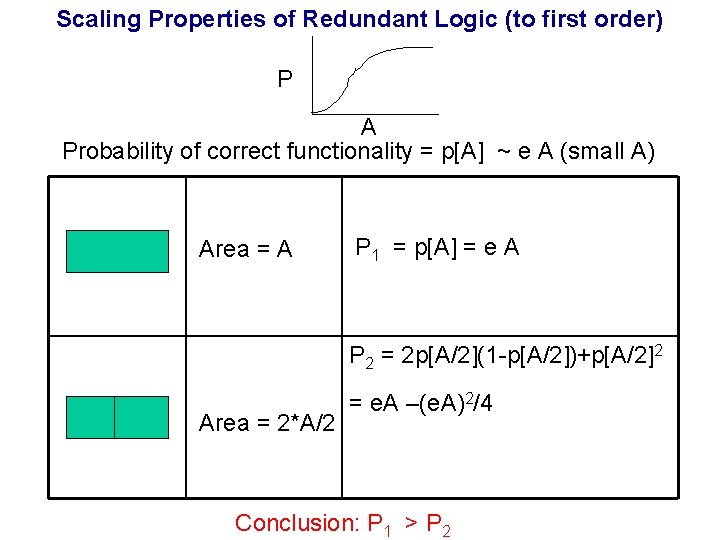
Designing Life Fault Tolerant Redundancy Error Correcting Fault Tolerant Other Coding (e. g. Parity) Error Correcting
![Designing Life I Fault Tolerant Redundancy Gene 1 Gene 2 Gene 3 Designing Life I] Fault Tolerant Redundancy Gene 1 Gene 2 Gene 3](https://slidetodoc.com/presentation_image_h2/61179f5eed7e860bea77885f2f547fe3/image-5.jpg)
Designing Life I] Fault Tolerant Redundancy Gene 1 Gene 2 Gene 3

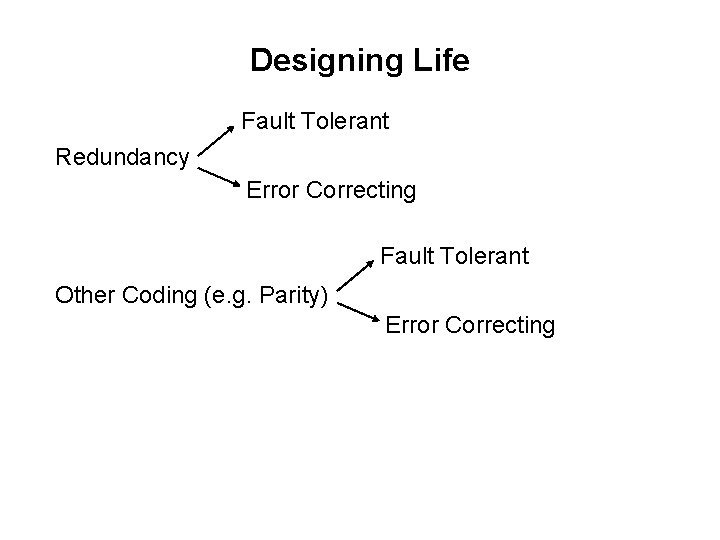
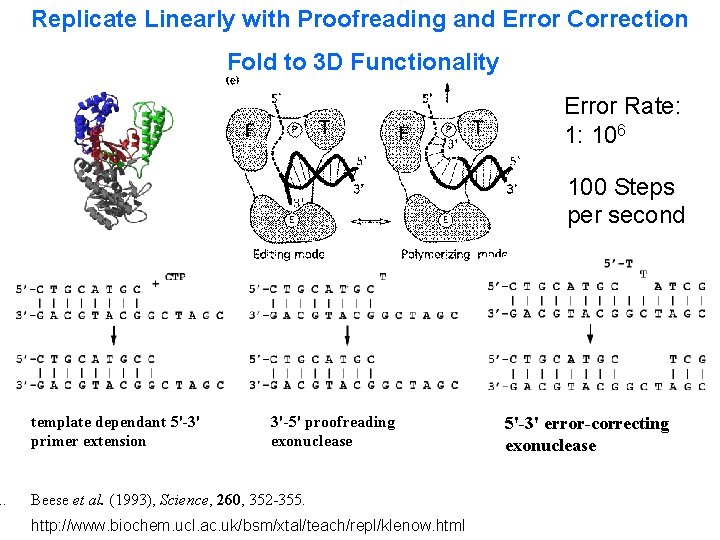
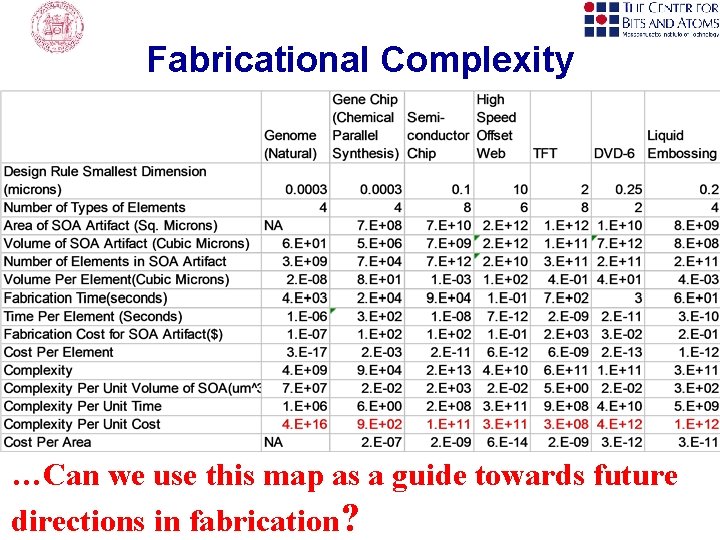
1. Replicate Linearly with Proofreading and Error Correction Fold to 3 D Functionality Error Rate: 1: 106 100 Steps per second template dependant 5'-3' primer extension 3'-5' proofreading exonuclease Beese et al. (1993), Science, 260, 352 -355. http: //www. biochem. ucl. ac. uk/bsm/xtal/teach/repl/klenow. html 5'-3' error-correcting exonuclease

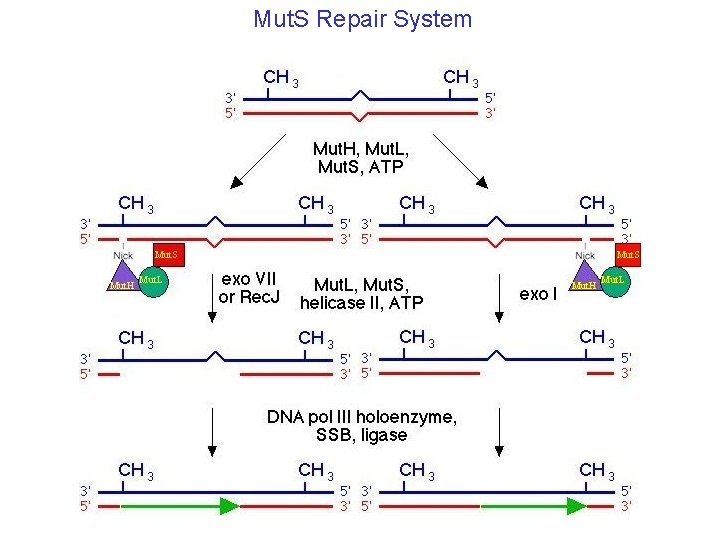
Mut. S Repair System
![Approach 1 b Redundant Genomes Deinococcus radiodurans 3 2 Mb 4 10 Copies of Approach 1 b] Redundant Genomes Deinococcus radiodurans (3. 2 Mb, 4 -10 Copies of](https://slidetodoc.com/presentation_image_h2/61179f5eed7e860bea77885f2f547fe3/image-8.jpg)
Approach 1 b] Redundant Genomes Deinococcus radiodurans (3. 2 Mb, 4 -10 Copies of Genome ) [Nature Biotechnology 18, 85 -90 (January 2000)] D. radiodurans: E. coli: Uniformed Services University of the Health 1. 7 Million Rads (17 k. Gy) – 200 DS breaks 25 Thousand Rads – 2 or 3 DS breaks http: //www. ornl. gov/hgmis/publicat/microbial/image 3. html

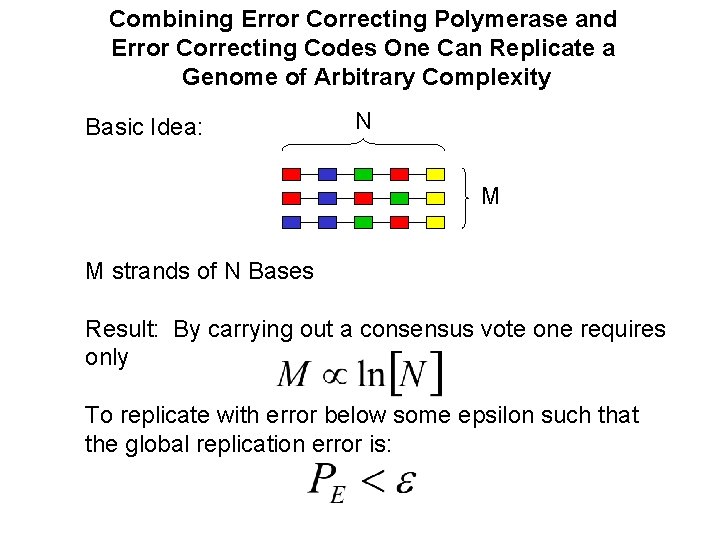
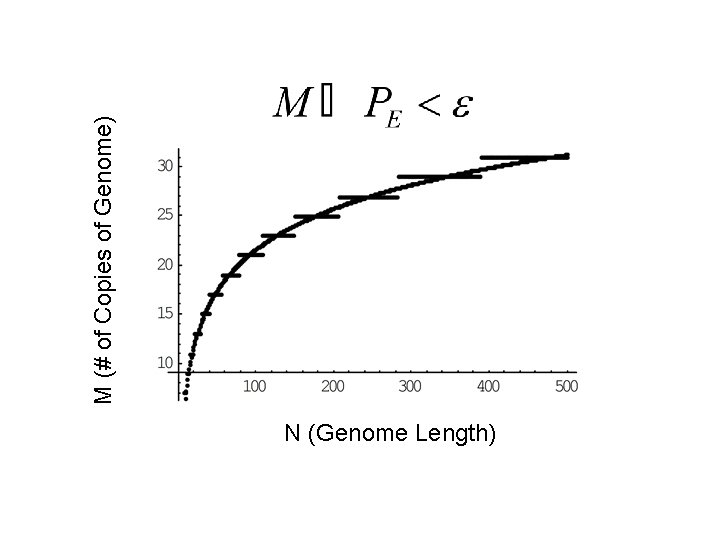
Combining Error Correcting Polymerase and Error Correcting Codes One Can Replicate a Genome of Arbitrary Complexity Basic Idea: N M M strands of N Bases Result: By carrying out a consensus vote one requires only To replicate with error below some epsilon such that the global replication error is:

N (Genome Length) M (# of Copies of Genome)
![II Coding m RNA Ribosome Amino Acid II] Coding m. RNA Ribosome Amino Acid](https://slidetodoc.com/presentation_image_h2/61179f5eed7e860bea77885f2f547fe3/image-11.jpg)
II] Coding m. RNA Ribosome Amino Acid

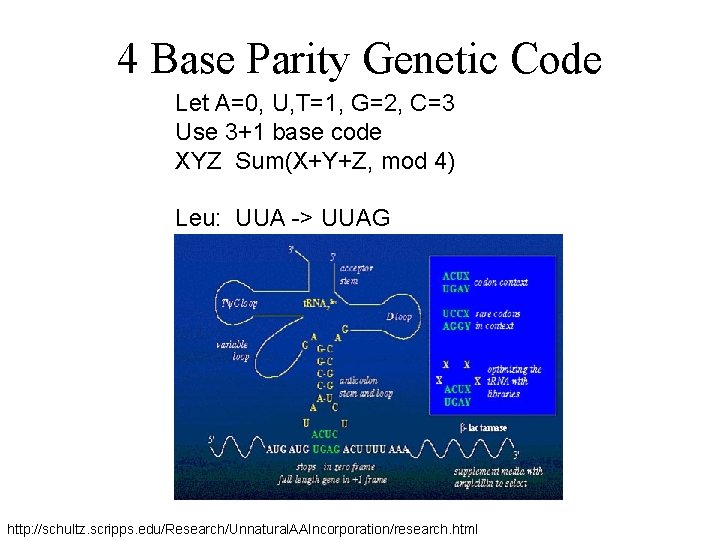
4 Base Parity Genetic Code Let A=0, U, T=1, G=2, C=3 Use 3+1 base code XYZ Sum(X+Y+Z, mod 4) Leu: UUA -> UUAG http: //schultz. scripps. edu/Research/Unnatural. AAIncorporation/research. html

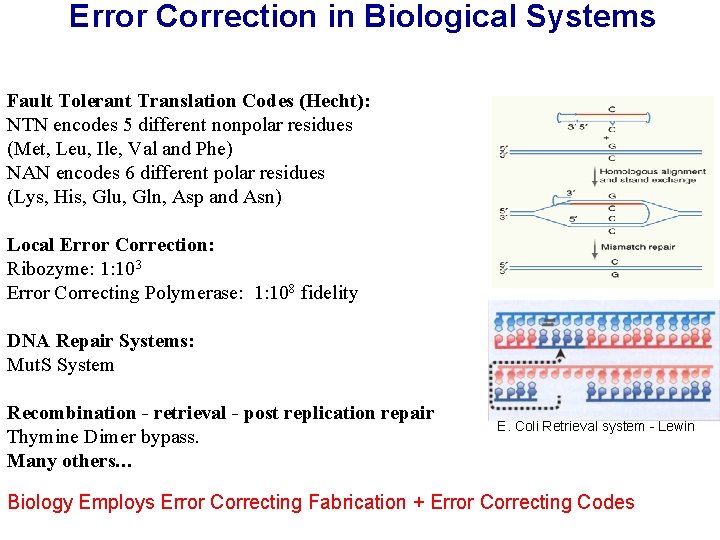
Error Correction in Biological Systems Fault Tolerant Translation Codes (Hecht): NTN encodes 5 different nonpolar residues (Met, Leu, Ile, Val and Phe) NAN encodes 6 different polar residues (Lys, His, Glu, Gln, Asp and Asn) Local Error Correction: Ribozyme: 1: 103 Error Correcting Polymerase: 1: 108 fidelity DNA Repair Systems: Mut. S System Recombination - retrieval - post replication repair Thymine Dimer bypass. Many others… E. Coli Retrieval system - Lewin Biology Employs Error Correcting Fabrication + Error Correcting Codes
![Physics of Information Technology MIT Spring 2006 410 1 Von Neumann Mc Physics of Information Technology MIT – Spring 2006 4/10 1] Von Neumann / Mc.](https://slidetodoc.com/presentation_image_h2/61179f5eed7e860bea77885f2f547fe3/image-14.jpg)
Physics of Information Technology MIT – Spring 2006 4/10 1] Von Neumann / Mc. Cullough/Winograd/Cowan Threshold Theorem and Fault Tolerant Chips 2] Simple Proofs in CMOS Scaling and Fault Tolerance 3] Fault Tolerant Self Replicating Systems 4] Fault Tolerant Codes in Biology 4/24 1]Introduction of the concept of Fabricational Complexity 2]Examples, numbers and mechanisms from native biology: error correcting polymerase and comparison to best current chemical synthesis using protection group (~feedforward) chemistry. 3]Examples from our error correcting de novo DNA synthesis (with hopefully a demo from our DNA synth simulator) 4]Error correcting chip synthesis 5]Saul's self replicating system with and without error correction

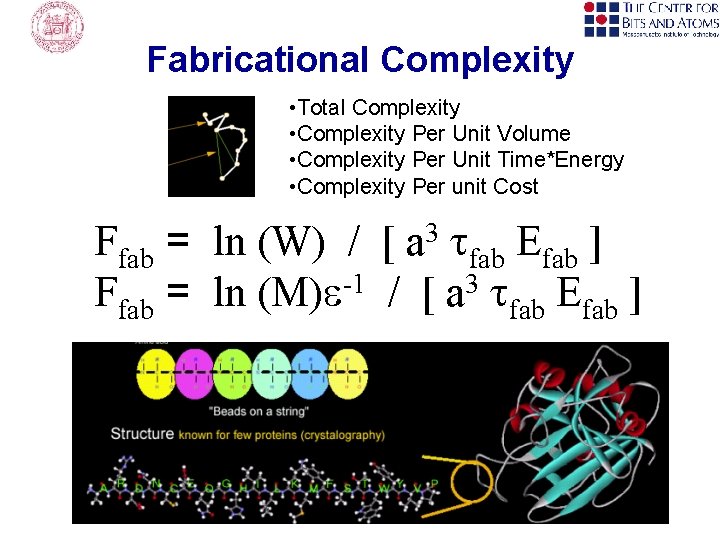
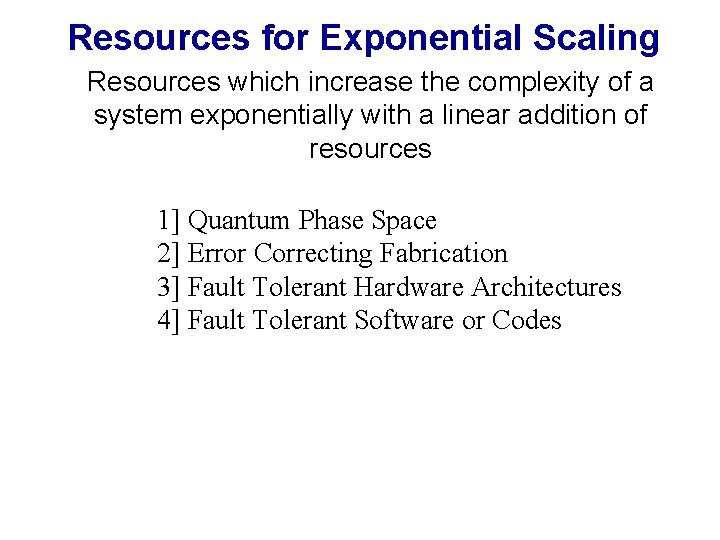
Fabricational Complexity • Total Complexity • Complexity Per Unit Volume • Complexity Per Unit Time*Energy • Complexity Per unit Cost Ffab = ln (W) / [ a 3 tfab Efab ] Ffab = ln (M)e-1 / [ a 3 tfab Efab ]

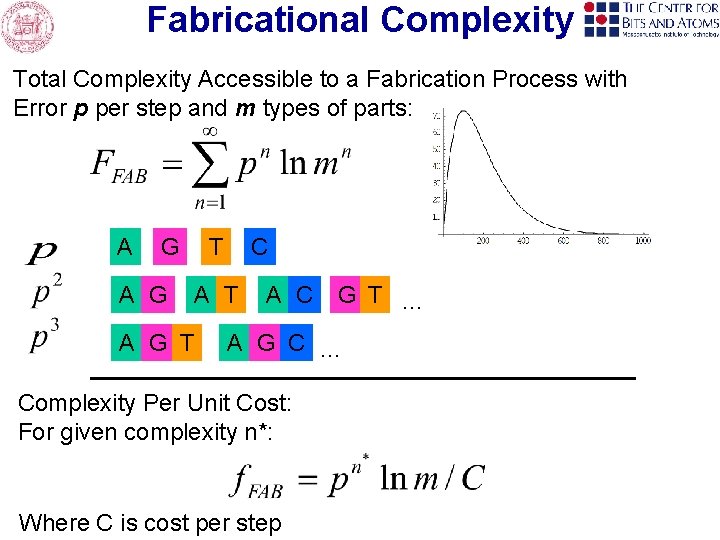
Fabricational Complexity Total Complexity Accessible to a Fabrication Process with Error p per step and m types of parts: A G T C A T A G T A C G T … A G C … Complexity Per Unit Cost: For given complexity n*: Where C is cost per step

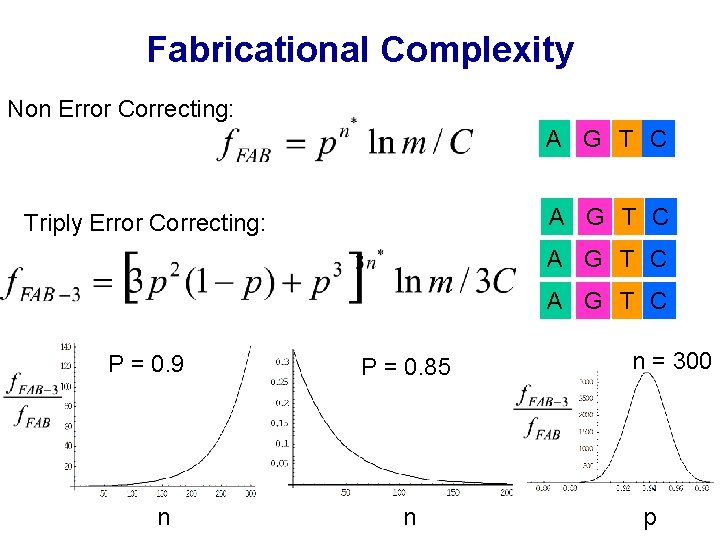
Fabricational Complexity Non Error Correcting: A G T C Triply Error Correcting: A G T C P = 0. 9 n P = 0. 85 n n = 300 p

Resources for Exponential Scaling Resources which increase the complexity of a system exponentially with a linear addition of resources 1] Quantum Phase Space 2] Error Correcting Fabrication 3] Fault Tolerant Hardware Architectures 4] Fault Tolerant Software or Codes

Fabricational Complexity …Can we use this map as a guide towards future directions in fabrication?

1. Replicate Linearly with Proofreading and Error Correction Fold to 3 D Functionality Error Rate: 1: 106 100 Steps per second template dependant 5'-3' primer extension 3'-5' proofreading exonuclease Beese et al. (1993), Science, 260, 352 -355. http: //www. biochem. ucl. ac. uk/bsm/xtal/teach/repl/klenow. html 5'-3' error-correcting exonuclease

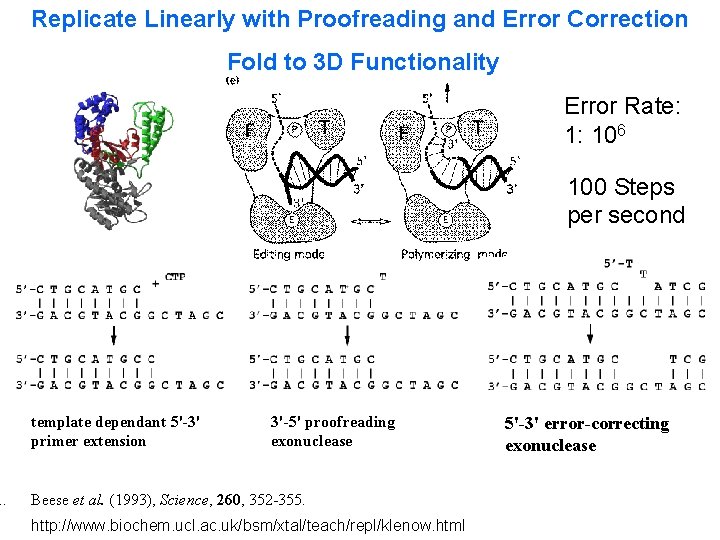
DNA Synthesis Caruthers Synthesis Error Rate: 1: 102 300 Seconds Per step http: //www. med. upenn. edu/naf /services/catalog 99. pdf

Avogadro Scale Engineering Molecular Machine (Jacobson) Group – MIT - May, 2005

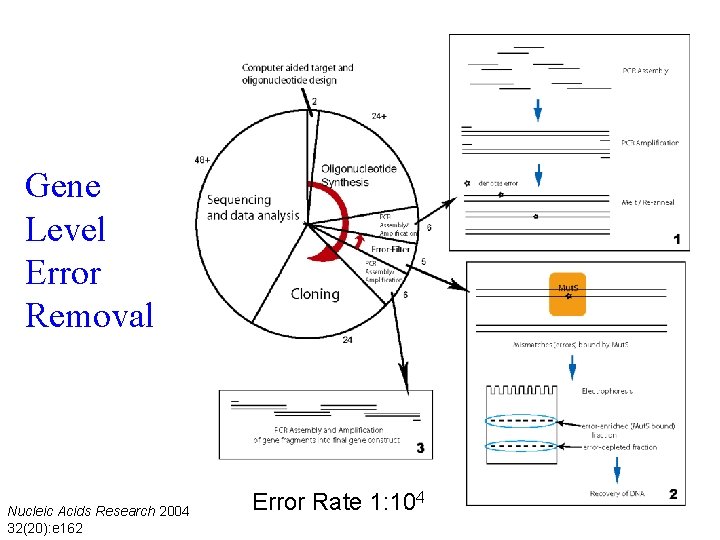
Gene Level Error Removal Nucleic Acids Research 2004 32(20): e 162 Error Rate 1: 104

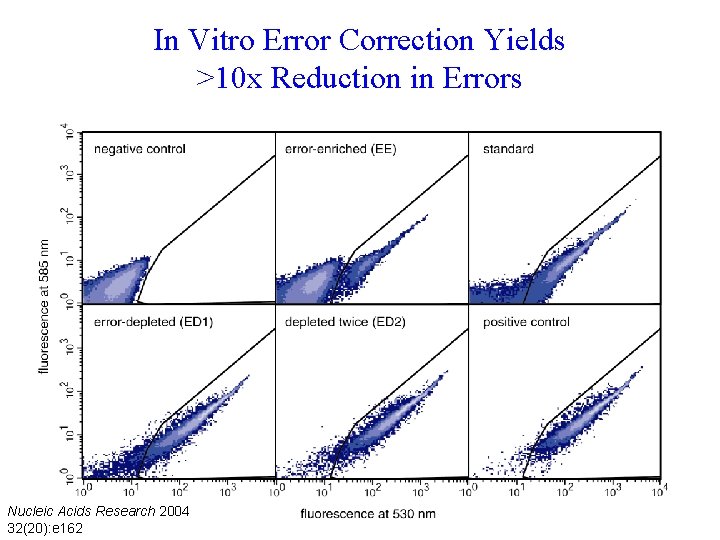
In Vitro Error Correction Yields >10 x Reduction in Errors Nucleic Acids Research 2004 32(20): e 162

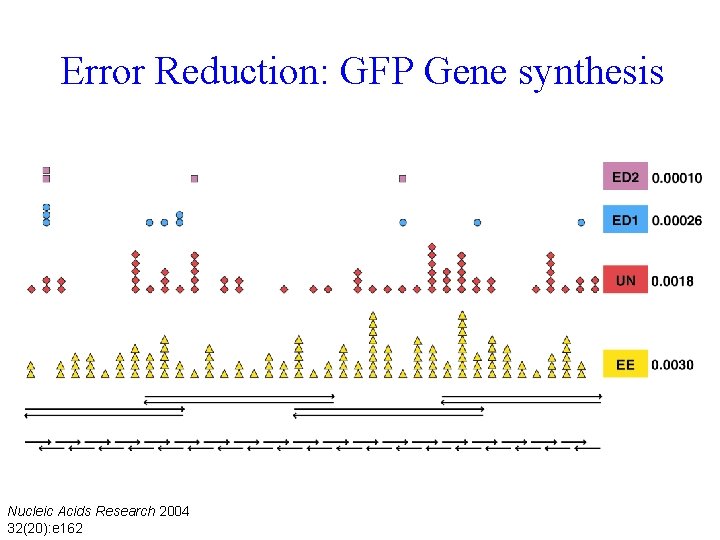
Error Reduction: GFP Gene synthesis Nucleic Acids Research 2004 32(20): e 162

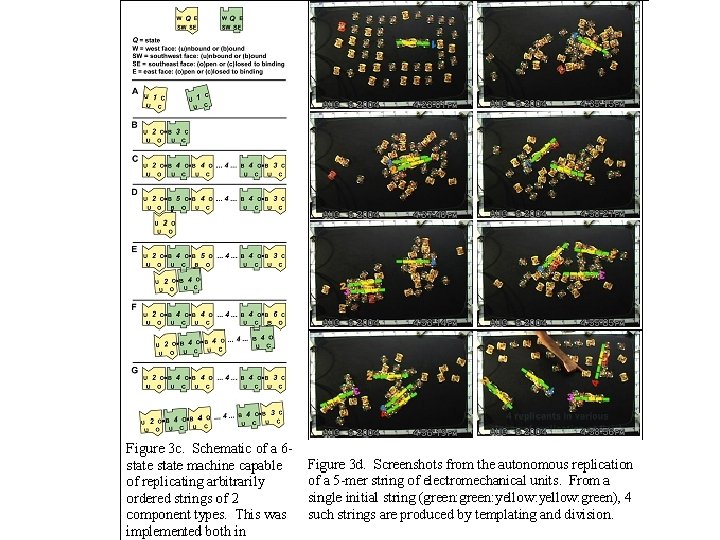
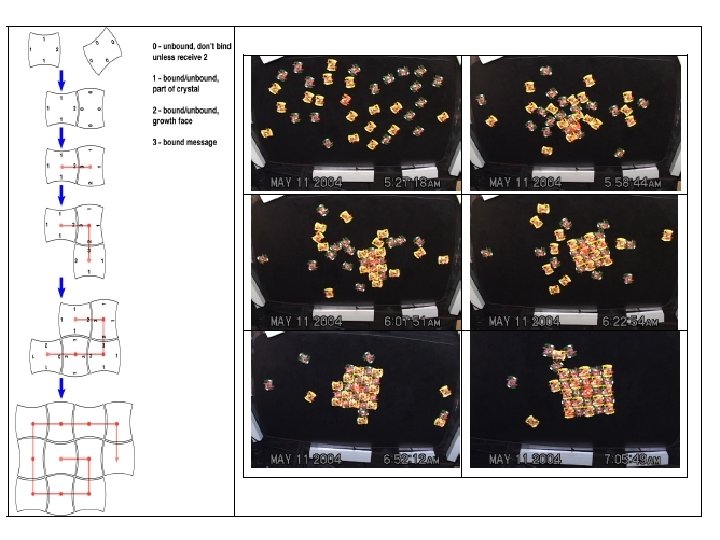
Autonomous self replicating machines from random building blocks



![HOMEWORK DUE 5106 1 Consider biological cells which are able to copy their HOMEWORK – DUE 5/1/06 1] Consider biological cells which are able to copy their](https://slidetodoc.com/presentation_image_h2/61179f5eed7e860bea77885f2f547fe3/image-30.jpg)
HOMEWORK – DUE 5/1/06 1] Consider biological cells which are able to copy their genome using appropriate pieces of molecular machinery (e. g. polymerase). Assume that the total probability of correctly copying each nucleotide is p=. 999 per nucleotide. Calculate the Total Fabrication Complexity accessible to this system assuming that there are 4 types of nucleotides (i. e. A, G, C, T). Now assume that we have created a new type of cell which has a genome possessing six different types of nucleotides (i. e. A, G, C, T, X, Y). If we assume that we wish to keep the total Fabricational Complexity the same what must the probability per nucleotide addition, p, now be? 2] Consider now the fabricational complexity per unit cost f. Calculate threshold probability p for which it is advantageous to use a redundant error correction scheme (such as trible redundancy) and majority voting than no error correction. Into which regime does biology fall?