Automatic Segmentation of Neonatal Brain MRI Marcel Prastawa
Automatic Segmentation of Neonatal Brain MRI Marcel Prastawa 1, John Gilmore 2, Weili Lin 3, Guido Gerig 1, 2 University of North Carolina at Chapel Hill 1 Department of Computer Science 2 Department of Psychiatry 3 Department of Radiology Partially supported by NIH Conte Center MH 064065 and NIH-NIBIB R 01 EB 000219 September 27, 2004 1 / 18 MIDAG@UNC
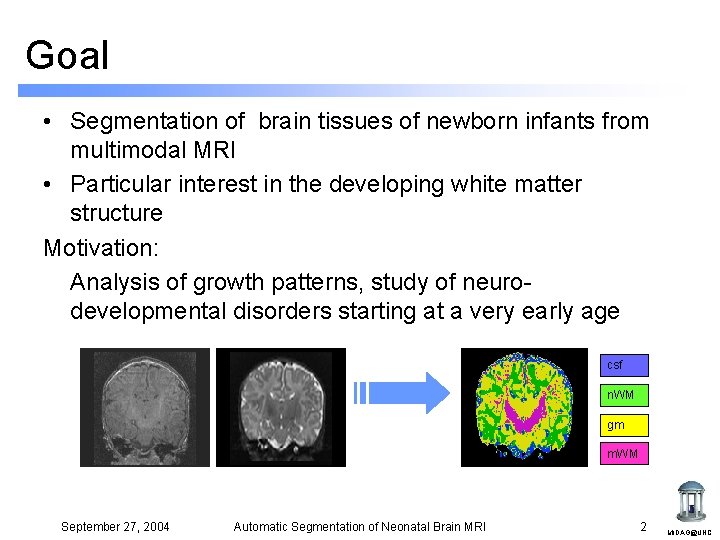
Goal • Segmentation of brain tissues of newborn infants from multimodal MRI • Particular interest in the developing white matter structure Motivation: Analysis of growth patterns, study of neurodevelopmental disorders starting at a very early age csf n. WM gm m. WM September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 2 MIDAG@UNC
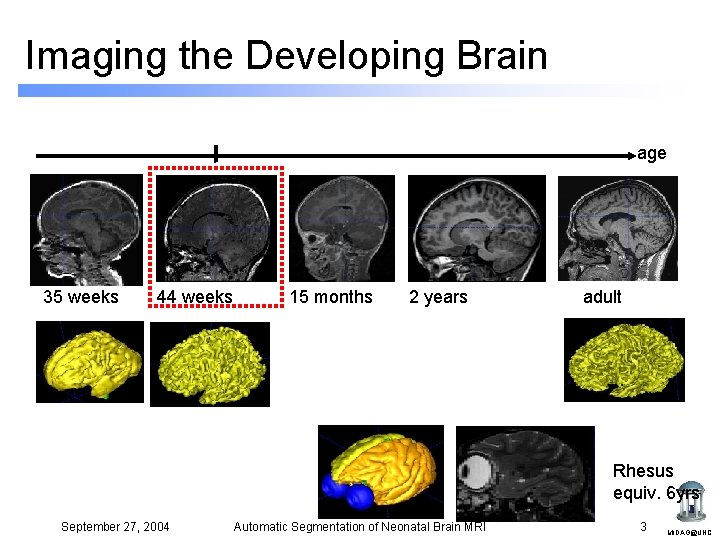
Imaging the Developing Brain age 35 weeks 44 weeks 15 months 2 years adult Rhesus equiv. 6 yrs September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 3 MIDAG@UNC
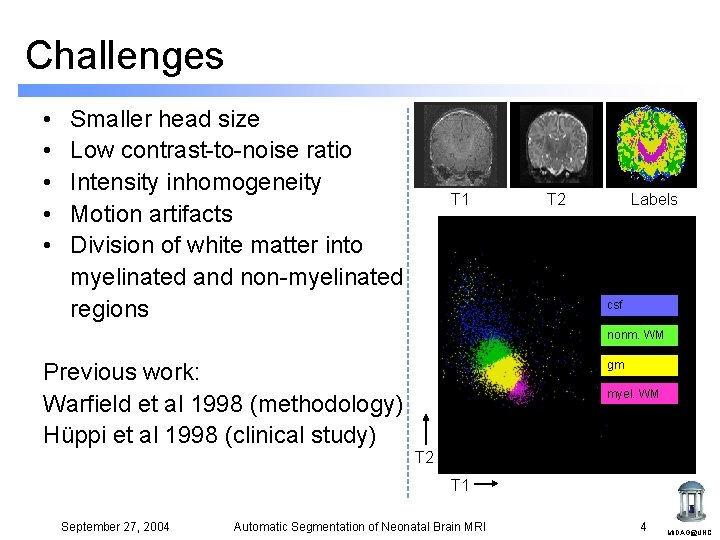
Challenges • • • Smaller head size Low contrast-to-noise ratio Intensity inhomogeneity Motion artifacts Division of white matter into myelinated and non-myelinated regions T 1 T 2 Labels csf nonm. WM Previous work: Warfield et al 1998 (methodology) Hüppi et al 1998 (clinical study) gm myel. WM T 2 T 1 September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 4 MIDAG@UNC
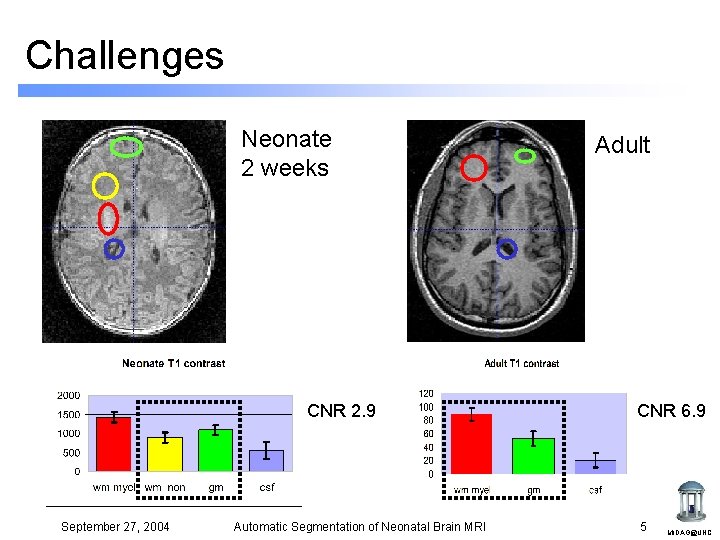
Challenges Neonate 2 weeks CNR 2. 9 September 27, 2004 Automatic Segmentation of Neonatal Brain MRI Adult CNR 6. 9 5 MIDAG@UNC
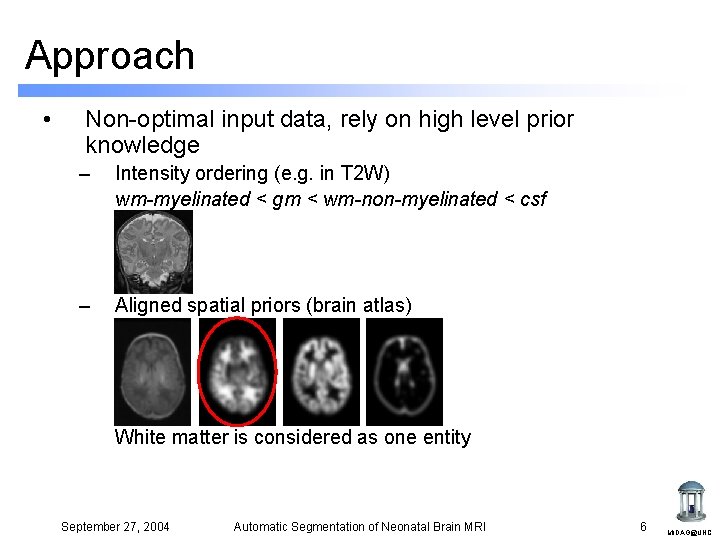
Approach • Non-optimal input data, rely on high level prior knowledge – Intensity ordering (e. g. in T 2 W) wm-myelinated < gm < wm-non-myelinated < csf – Aligned spatial priors (brain atlas) White matter is considered as one entity September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 6 MIDAG@UNC
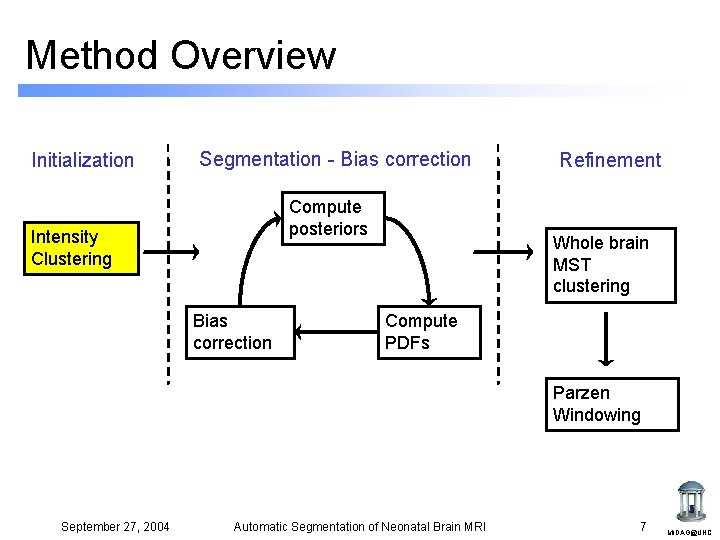
Method Overview Initialization Segmentation - Bias correction Compute posteriors Intensity Clustering Bias correction Refinement Whole brain MST clustering Compute PDFs Parzen Windowing September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 7 MIDAG@UNC
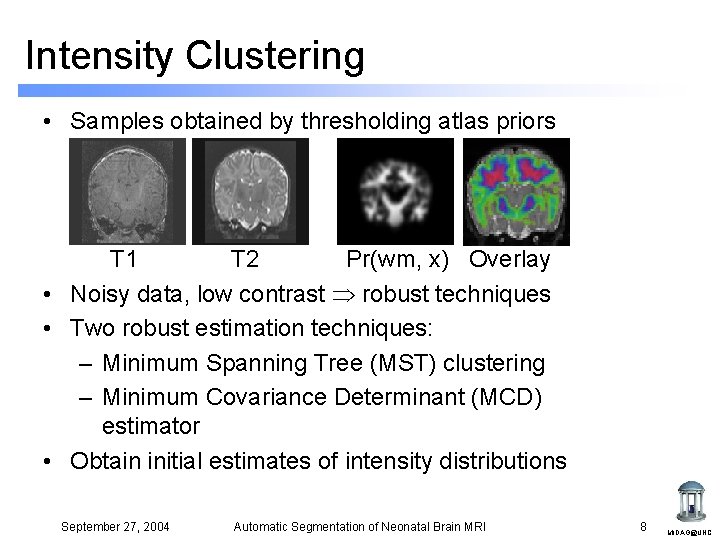
Intensity Clustering • Samples obtained by thresholding atlas priors T 1 T 2 Pr(wm, x) Overlay • Noisy data, low contrast robust techniques • Two robust estimation techniques: – Minimum Spanning Tree (MST) clustering – Minimum Covariance Determinant (MCD) estimator • Obtain initial estimates of intensity distributions September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 8 MIDAG@UNC
![Minimum Spanning Tree Clustering [Cocosco et al 2003] • Break long edges in MST, Minimum Spanning Tree Clustering [Cocosco et al 2003] • Break long edges in MST,](http://slidetodoc.com/presentation_image/3f0e00b66ab3607fc1573367db4f8682/image-9.jpg)
Minimum Spanning Tree Clustering [Cocosco et al 2003] • Break long edges in MST, example: Detect multiple clusters while pruning outliers • Iterative process, stops when cluster feature locations are in the desired order • “Feature location” = summary value of cluster intensities September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 9 MIDAG@UNC
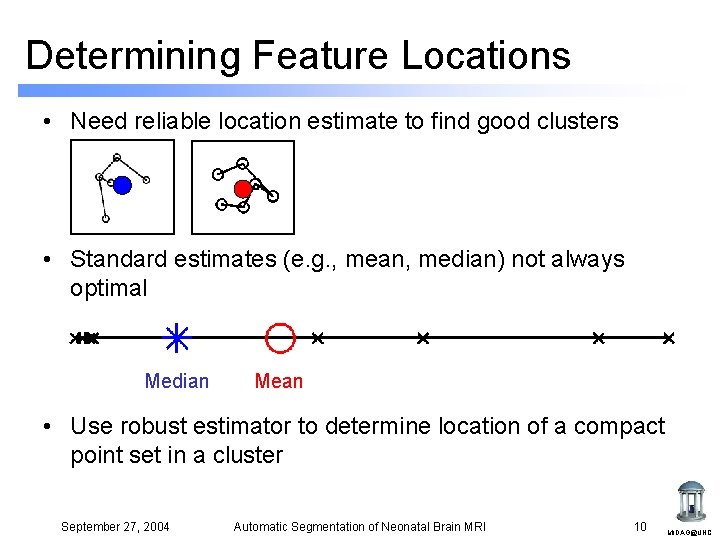
Determining Feature Locations • Need reliable location estimate to find good clusters • Standard estimates (e. g. , mean, median) not always optimal Median Mean • Use robust estimator to determine location of a compact point set in a cluster September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 10 MIDAG@UNC
![Minimum Covariance Determinant [Rousseeuw et al 1999] • Feature location of MST clusters to Minimum Covariance Determinant [Rousseeuw et al 1999] • Feature location of MST clusters to](http://slidetodoc.com/presentation_image/3f0e00b66ab3607fc1573367db4f8682/image-11.jpg)
Minimum Covariance Determinant [Rousseeuw et al 1999] • Feature location of MST clusters to determine ordering? • Smallest ellipsoid that covers at least half the data • MCD gives robust location estimate • Example: O = points used for estimation X = other data points September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 11 MIDAG@UNC
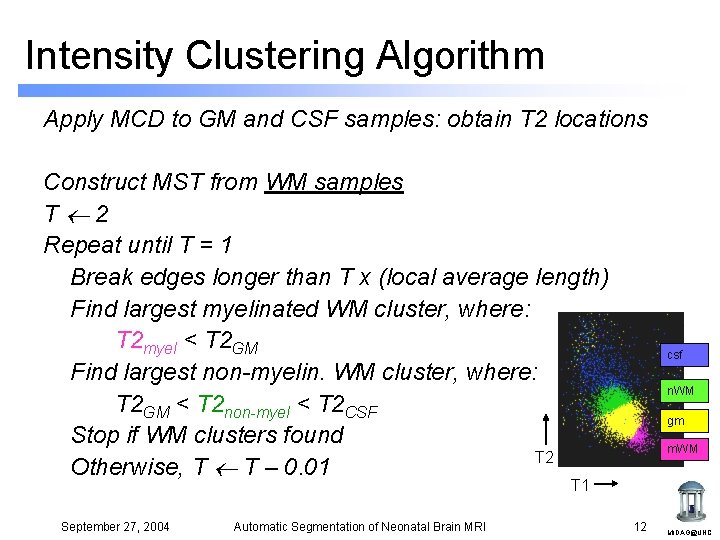
Intensity Clustering Algorithm Apply MCD to GM and CSF samples: obtain T 2 locations Construct MST from WM samples T 2 Repeat until T = 1 Break edges longer than T x (local average length) Find largest myelinated WM cluster, where: T 2 myel < T 2 GM Find largest non-myelin. WM cluster, where: T 2 GM < T 2 non-myel < T 2 CSF Stop if WM clusters found T 2 Otherwise, T T – 0. 01 csf n. WM gm m. WM T 1 September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 12 MIDAG@UNC
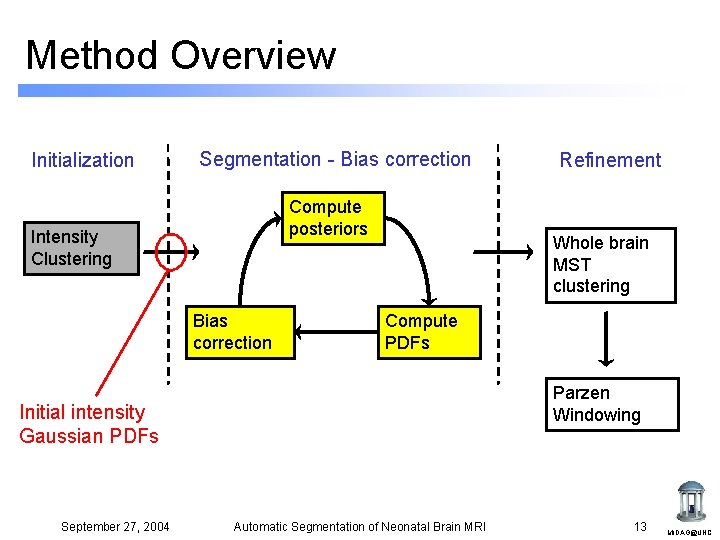
Method Overview Initialization Segmentation - Bias correction Compute posteriors Intensity Clustering Bias correction Whole brain MST clustering Compute PDFs Parzen Windowing Initial intensity Gaussian PDFs September 27, 2004 Refinement Automatic Segmentation of Neonatal Brain MRI 13 MIDAG@UNC
![Bias Correction [Wells et al 1996, van Leemput et al 1999] • “Bias” = Bias Correction [Wells et al 1996, van Leemput et al 1999] • “Bias” =](http://slidetodoc.com/presentation_image/3f0e00b66ab3607fc1573367db4f8682/image-14.jpg)
Bias Correction [Wells et al 1996, van Leemput et al 1999] • “Bias” = RF inhomogeneity and biology • Images low contrast, histogram is smooth • Use spatial context, bias is log-difference of input intensities and reconstructed “flat” image bias biology • Fit polynomial to the bias field (weighted least squares) • Interleaves segmentation and bias correction Compute posteriors Bias correction September 27, 2004 Gaussian intensity PDFs Compute PDFs Automatic Segmentation of Neonatal Brain MRI 14 MIDAG@UNC
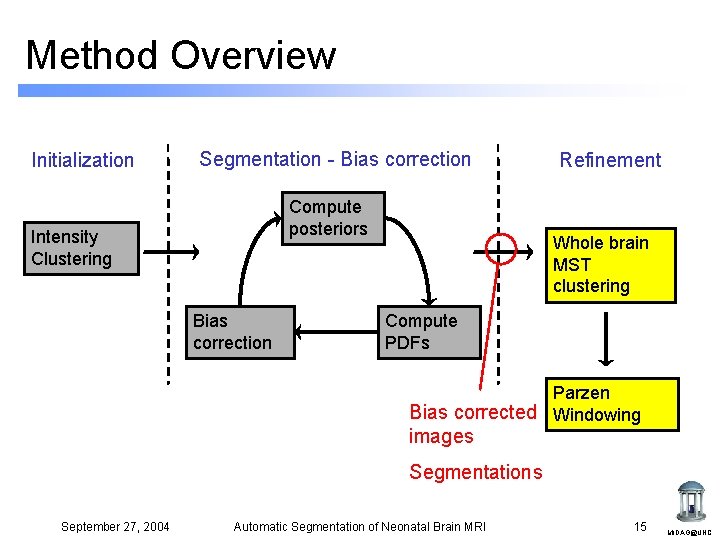
Method Overview Initialization Segmentation - Bias correction Compute posteriors Intensity Clustering Bias correction Refinement Whole brain MST clustering Compute PDFs Parzen Bias corrected Windowing images Segmentations September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 15 MIDAG@UNC
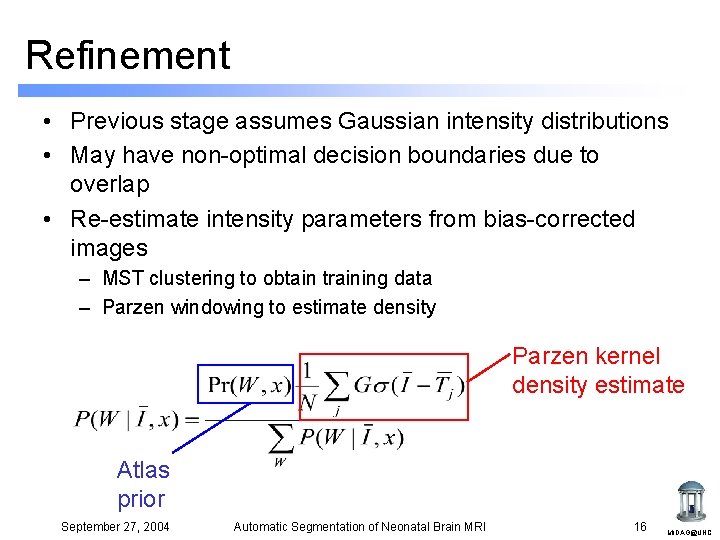
Refinement • Previous stage assumes Gaussian intensity distributions • May have non-optimal decision boundaries due to overlap • Re-estimate intensity parameters from bias-corrected images – MST clustering to obtain training data – Parzen windowing to estimate density Parzen kernel density estimate Atlas prior September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 16 MIDAG@UNC
![Results [1/2] • UNC Radiology Weili Lin (Siemens 3 T head-only) • UNC-0094 T Results [1/2] • UNC Radiology Weili Lin (Siemens 3 T head-only) • UNC-0094 T](http://slidetodoc.com/presentation_image/3f0e00b66ab3607fc1573367db4f8682/image-17.jpg)
Results [1/2] • UNC Radiology Weili Lin (Siemens 3 T head-only) • UNC-0094 T 1 T 2 Classification 3 D • UNC-0096 T 1 September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 17 MIDAG@UNC
![Results [2/2] • Provided by Petra Hüppi (Geneva, Philips 1. 5 T) • Geneva-001 Results [2/2] • Provided by Petra Hüppi (Geneva, Philips 1. 5 T) • Geneva-001](http://slidetodoc.com/presentation_image/3f0e00b66ab3607fc1573367db4f8682/image-18.jpg)
Results [2/2] • Provided by Petra Hüppi (Geneva, Philips 1. 5 T) • Geneva-001 T 2 Classification 3 D • Geneva-002 T 1 September 27, 2004 T 2 Classification 3 D Automatic Segmentation of Neonatal Brain MRI 18 MIDAG@UNC
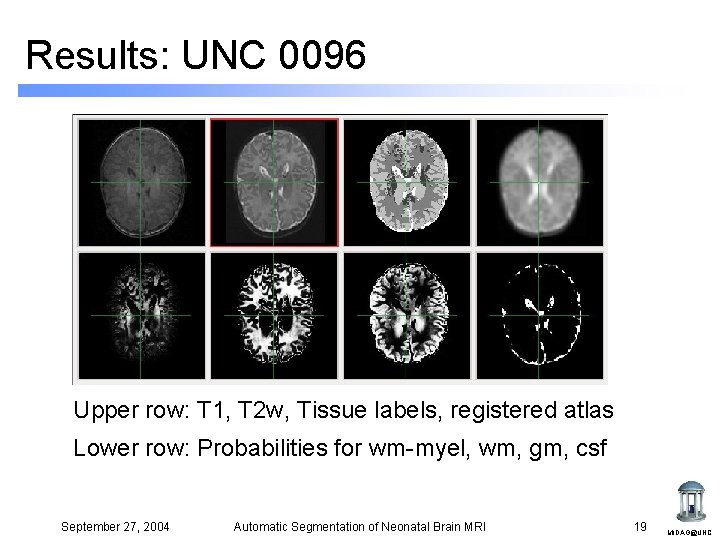
Results: UNC 0096 Upper row: T 1, T 2 w, Tissue labels, registered atlas Lower row: Probabilities for wm-myel, wm, gm, csf September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 19 MIDAG@UNC
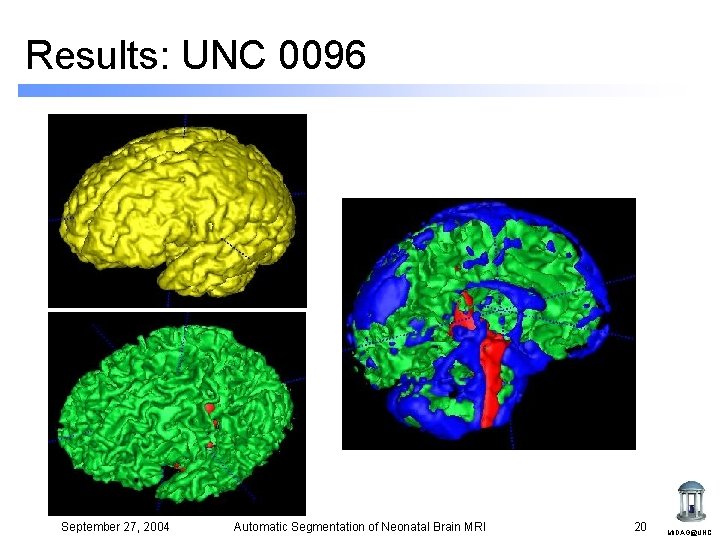
Results: UNC 0096 September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 20 MIDAG@UNC
Summary • Automatic brain tissue segmentation of neonatal MRI • Detects white matter as myelinated and non-myelinated structures • Makes use of prior knowledge: – Image intensity ordering – Spatial locations (probabilistic atlas prior) • To be used in two large UNC neonatal MRI studies – Silvio Conte Center: 125 neonates at risk – Neonate Twin study (heritability) • Current focus: Validation September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 21 MIDAG@UNC
Acknowledgements • • Elizabeth Bullitt Petra Hüppi Koen van Leemput Insight Toolkit Community Neoseg v 1. 0 b September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 22 MIDAG@UNC
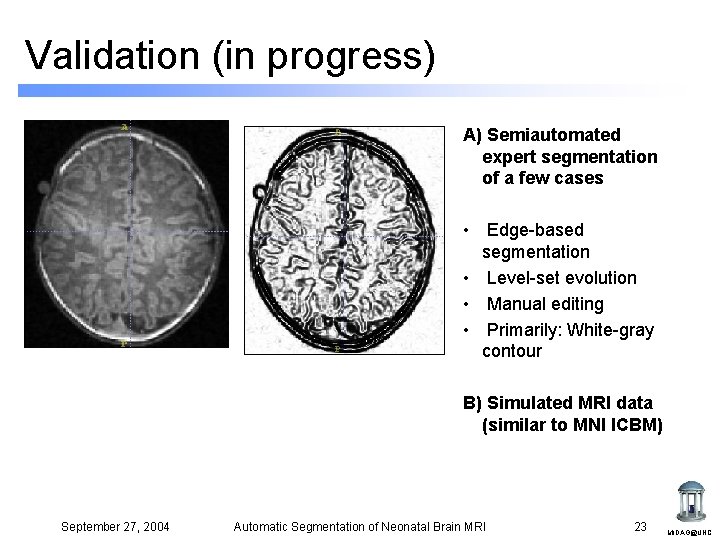
Validation (in progress) A) Semiautomated expert segmentation of a few cases • Edge-based segmentation • Level-set evolution • Manual editing • Primarily: White-gray contour B) Simulated MRI data (similar to MNI ICBM) September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 23 MIDAG@UNC
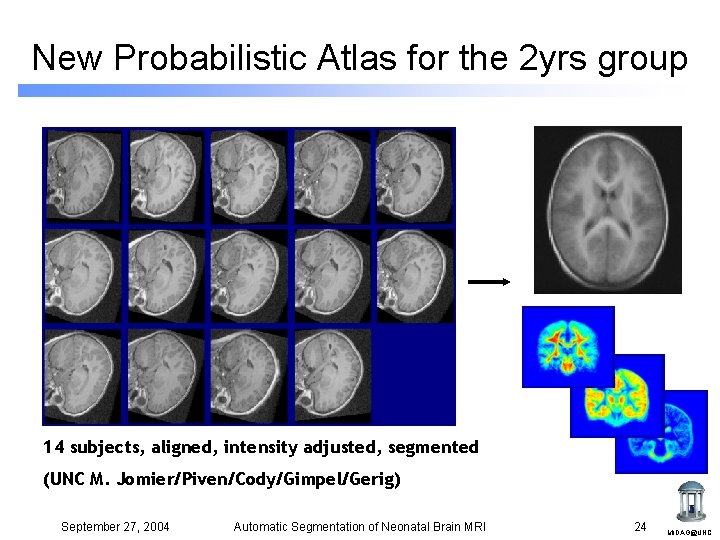
New Probabilistic Atlas for the 2 yrs group 14 subjects, aligned, intensity adjusted, segmented (UNC M. Jomier/Piven/Cody/Gimpel/Gerig) September 27, 2004 Automatic Segmentation of Neonatal Brain MRI 24 MIDAG@UNC
- Slides: 24