AUTOMATIC SEGMENTATION OF MR BRAIN IMAGES WITH A
AUTOMATIC SEGMENTATION OF MR BRAIN IMAGES WITH A CONVOLUTIONAL NEURAL NETWORK Pim Moeskops, Max A. Viergever, Adriënne M. Mendrik, Linda S. de Vries, Manon J. N. L. Benders, and Ivana Išgum IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 35, NO. 5, MAY 2016

BRAIN MRI SEGMENTATION Automatic MRI segmentation: • Used for measuring and visualizing the brain's anatomic structures • Analyzing brain changes • Delineating pathologic regions • Important for quantitative analysis in largescale studies with images acquired at all ages • Large scale manual annotation of 3 D MRI scans is not feasible
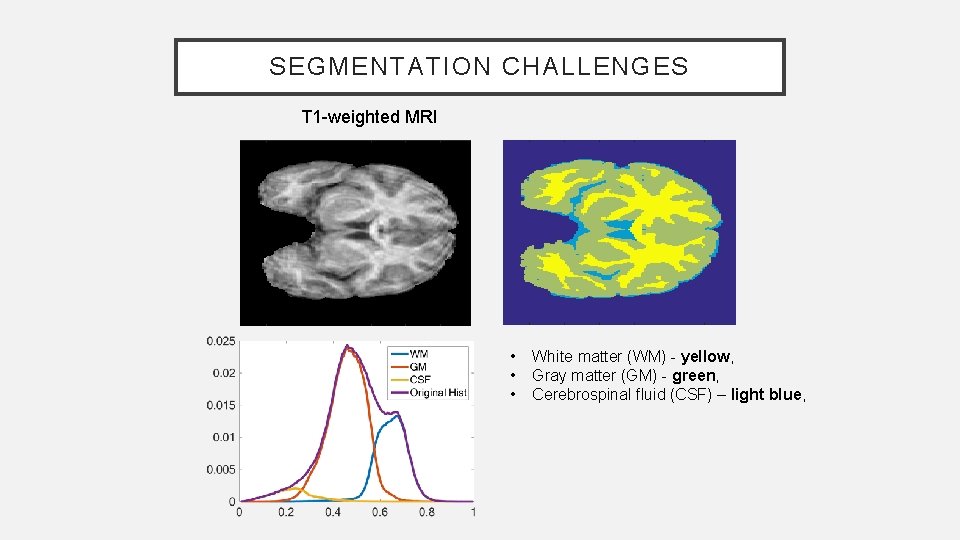
SEGMENTATION CHALLENGES T 1 -weighted MRI • • • White matter (WM) - yellow, Gray matter (GM) - green, Cerebrospinal fluid (CSF) – light blue,
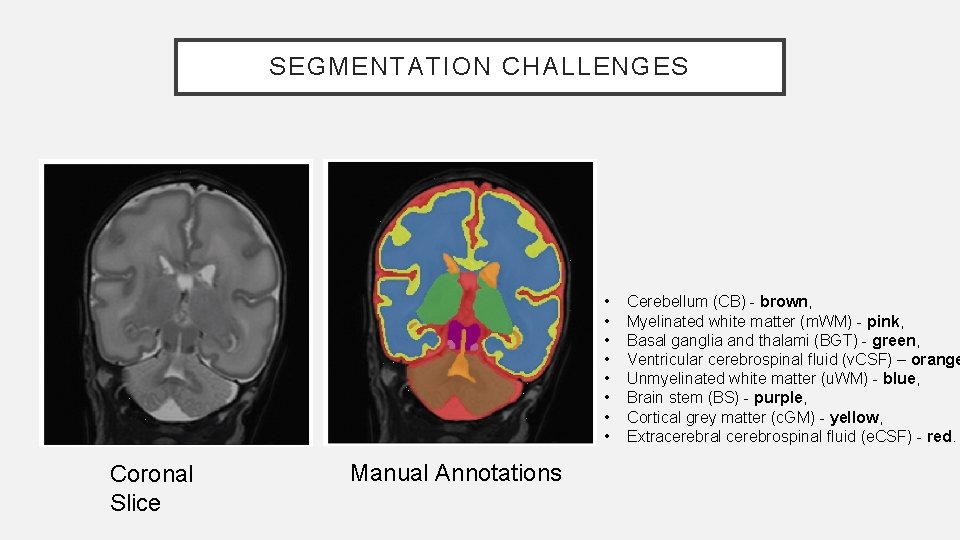
SEGMENTATION CHALLENGES • • Coronal Slice Manual Annotations Cerebellum (CB) - brown, Myelinated white matter (m. WM) - pink, Basal ganglia and thalami (BGT) - green, Ventricular cerebrospinal fluid (v. CSF) – orange Unmyelinated white matter (u. WM) - blue, Brain stem (BS) - purple, Cortical grey matter (c. GM) - yellow, Extracerebral cerebrospinal fluid (e. CSF) - red.

NETWORK STRUCTURE • Image patches of various scales are used to describe their central voxel. Patches are extracted only from imaging plane. • The larger scales keep implicit information about the voxel location in the scan • Smaller scales provide detailed information about the local neighborhood of the voxel
NETWORK STRUCTURE • A separate network branch for each of the patches scales • All network branches are connected to a single softmax output layer with one node for each class (including a background class). • Rectified linear units are used for all nodes • Drop-out is used on the fully connected layers • Mini-batch learning • Cross-entropy is used as cost function

PRE-PROCESSING • Images are bias corrected prior to segmentation, • Intensities are scaled to the 0 - 1023 range • Skull is removed K. V. Leemput, et al. “Automated model-based bias field correction of MR images of the brain”. IEEE TMI, 1999.
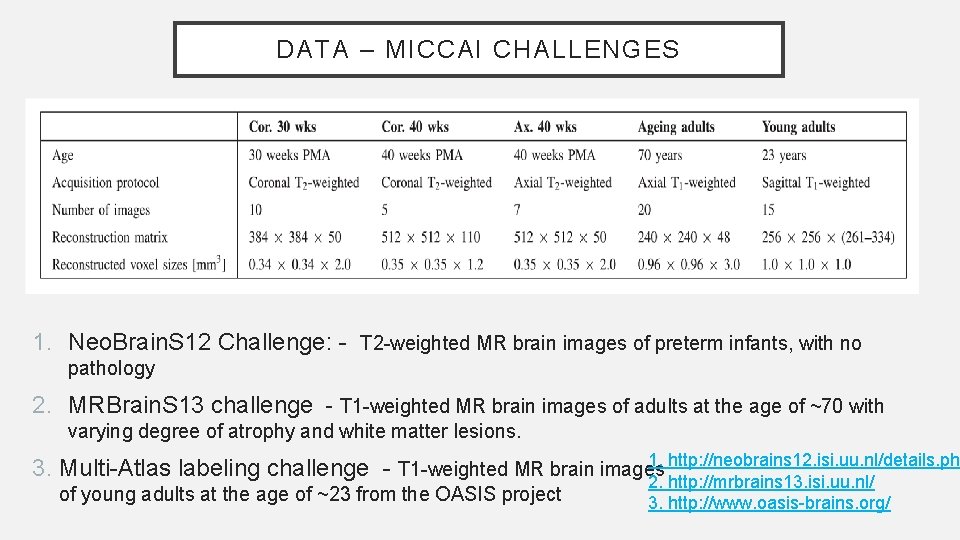
DATA – MICCAI CHALLENGES 1. Neo. Brain. S 12 Challenge: - T 2 -weighted MR brain images of preterm infants, with no pathology 2. MRBrain. S 13 challenge - T 1 -weighted MR brain images of adults at the age of ~70 with varying degree of atrophy and white matter lesions. 1. http: //neobrains 12. isi. uu. nl/details. php 3. Multi-Atlas labeling challenge - T 1 -weighted MR brain images 2. http: //mrbrains 13. isi. uu. nl/ of young adults at the age of ~23 from the OASIS project 3. http: //www. oasis-brains. org/
TRAINING AND PARAMETER TUNING • Parameter settings were defined in a preliminary leave-one-subject-out experiments with 5 of the images acquired at 30 weeks PMA. • Tissue classes consist of a very different number of voxels: i. e. 200 for m. WM and 400 K for u. WM in 30 weeks PMA. • Training samples includes a fixed number of samples per class from each training image. For smaller classes all samples are used. • A new random selection of samples is made in every epoch, and provided to the network in a randomized order • 10 epochs and 50 000 training samples per class from each image are used for training in all experiments
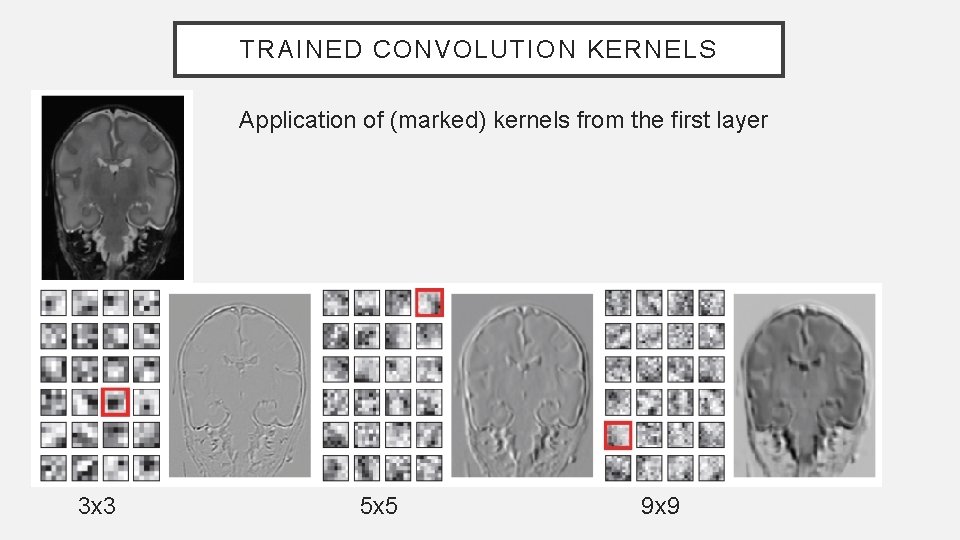
TRAINED CONVOLUTION KERNELS Application of (marked) kernels from the first layer 3 x 3 5 x 5 9 x 9
INFLUENCE OF MULTI-SCALE APPROACH (a) T 2 -weighted; (b) Manual Segmentation; (c) 21 x 21; (d) 51 x 51; (e) 75 x 75; (f) 3 scales combined
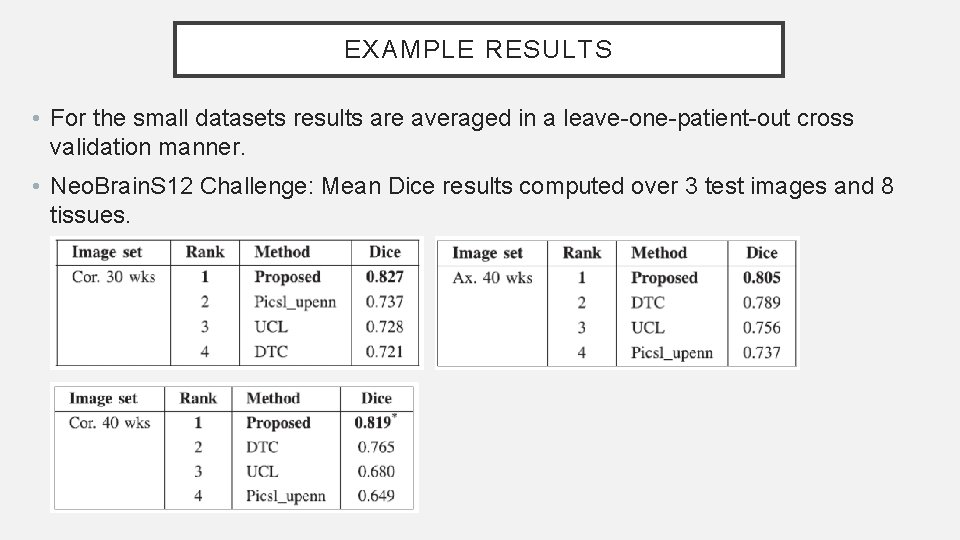
EXAMPLE RESULTS • For the small datasets results are averaged in a leave-one-patient-out cross validation manner. • Neo. Brain. S 12 Challenge: Mean Dice results computed over 3 test images and 8 tissues.
SUMMARY • A convolutional neural network that uses multiple patch sizes and multiple convolution kernel sizes to acquire multi-scale information about each voxel. • Training is performed from a single anatomical MR image • Provides accurate segmentation with spatial consistency • Results demonstrates robustness to differences in age and acquisition protocol.

DISCUSSION • CNNs is applied in a voxel classification task, thus many training samples are available. • Training using a single image, instead of relying on multiple acquisitions, allows omitting registration between images and thus precludes possible registration errors. • Training extracted from a limited number of training images. More training images could therefore increase the performance. • Including diverse training data from different data sets in a single training step
THANK YOU!
- Slides: 15