Automatic genomewide reconstruction of phylogenetic gene trees Ilan










- Slides: 60

Automatic genome-wide reconstruction of phylogenetic gene trees Ilan Wapinski, Avi Pfeffer, Nir Friedman, Aviv Regev Bioinformatics, Vol. 23, No. 13. (1 July 2007) Presented by : Hsin-Ta Wu CSCI-2950 C Topics in Computational Biology 2008. NOV. 4

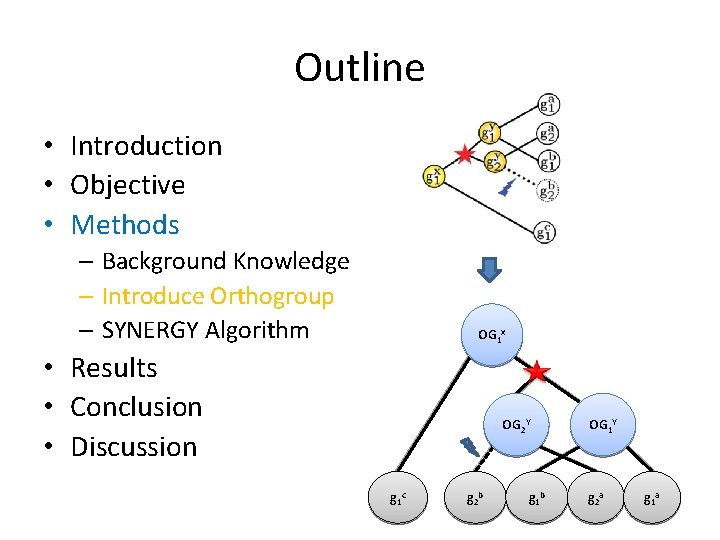
Outline • • • Introduction Objective Methods Results Conclusion Discussion

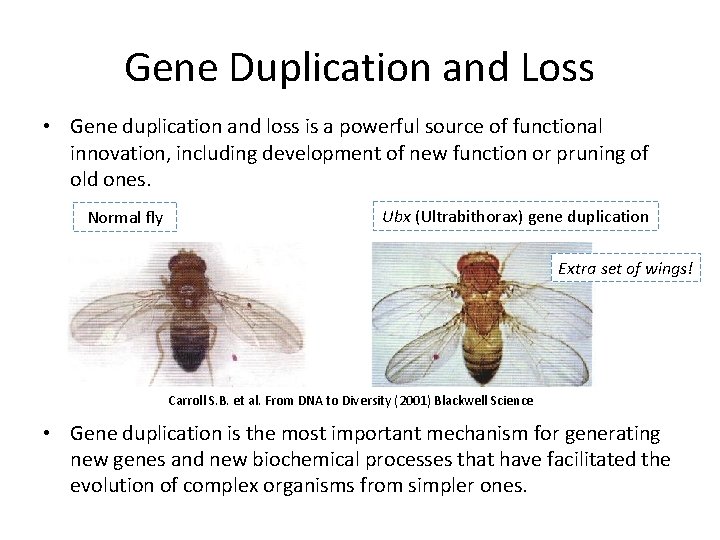
Gene Duplication and Loss • Gene duplication and loss is a powerful source of functional innovation, including development of new function or pruning of old ones. Normal fly Ubx (Ultrabithorax) gene duplication Extra set of wings! Carroll S. B. et al. From DNA to Diversity (2001) Blackwell Science • Gene duplication is the most important mechanism for generating new genes and new biochemical processes that have facilitated the evolution of complex organisms from simpler ones.

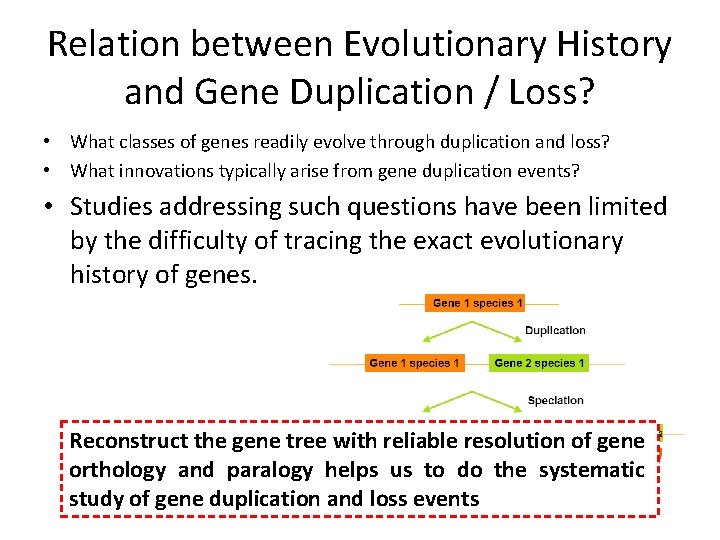
Relation between Evolutionary History and Gene Duplication / Loss? • What classes of genes readily evolve through duplication and loss? • What innovations typically arise from gene duplication events? • Studies addressing such questions have been limited by the difficulty of tracing the exact evolutionary history of genes. Reconstruct the gene tree with reliable resolution of gene orthology and paralogy helps us to do the systematic study of gene duplication and loss events

Outline • • • Introduction Objective Methods Results Conclusion Discussion

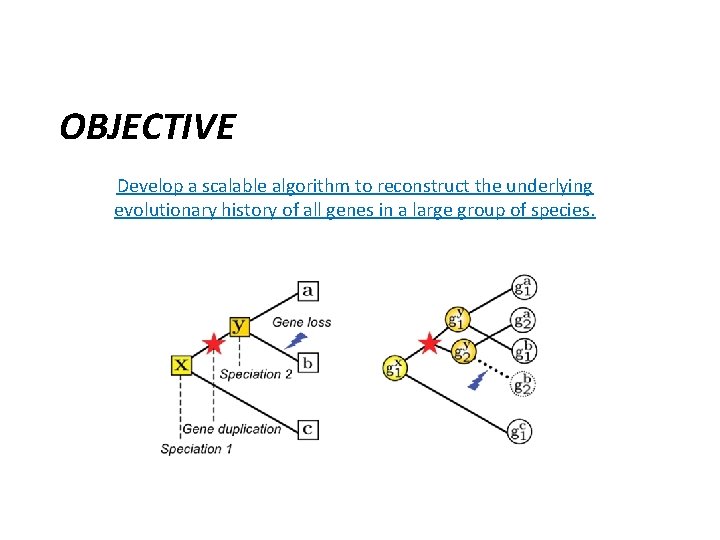
OBJECTIVE Develop a scalable algorithm to reconstruct the underlying evolutionary history of all genes in a large group of species.

Outline • Introduction • Objective • Methods – Background Knowledge – Introduce Orthogroup – SYNERGY Algorithm • Results • Conclusion • Discussion

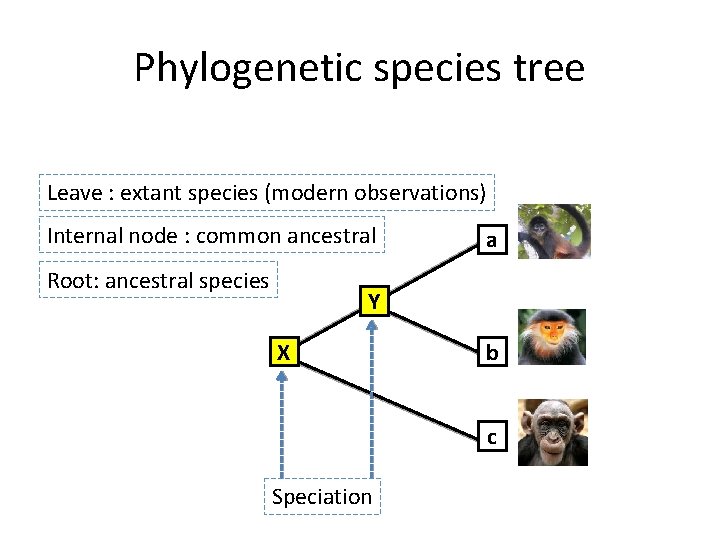
Phylogenetic species tree Leave : extant species (modern observations) Internal node : common ancestral Root: ancestral species a Y X b c Speciation

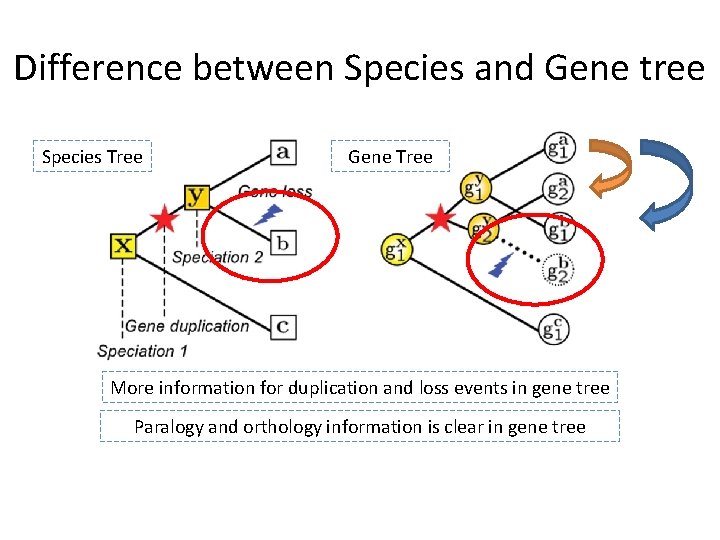
Difference between Species and Gene tree Species Tree Gene Tree More information for duplication and loss events in gene tree Paralogy and orthology information is clear in gene tree

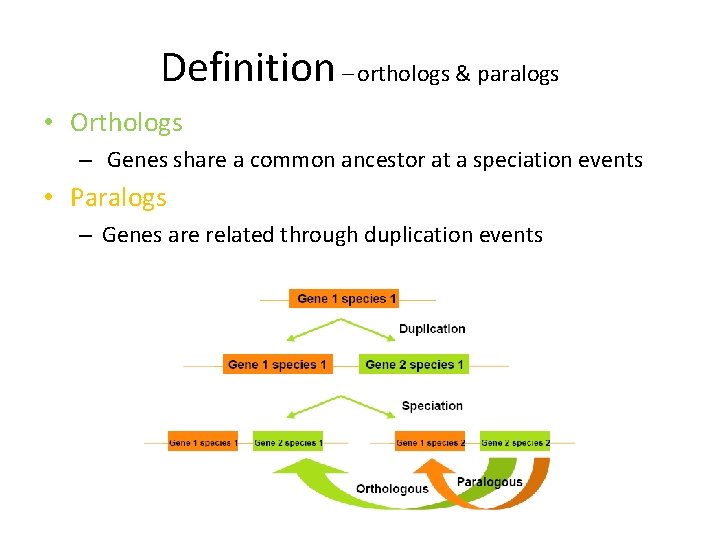
Definition – orthologs & paralogs • Orthologs – Genes share a common ancestor at a speciation events • Paralogs – Genes are related through duplication events

Outline • Introduction • Objective • Methods – Background Knowledge – Introduce Orthogroup – SYNERGY Algorithm OG 1 x • Results • Conclusion • Discussion OG 2 Y g 1 c g 2 b g 1 b OG 1 Y g 2 a g 1 a

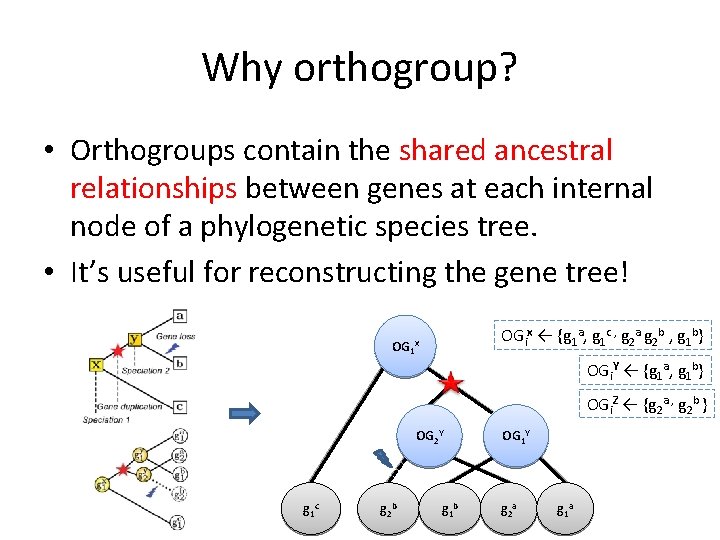
Why orthogroup? • Orthogroups contain the shared ancestral relationships between genes at each internal node of a phylogenetic species tree. • It’s useful for reconstructing the gene tree! OGix ← {g 1 a, g 1 c , g 2 a g 2 b , g 1 b} OG 1 x OGi. Y ← {g 1 a, g 1 b} OGi. Z ← {g 2 a , g 2 b } OG 2 Y g 1 c g 2 b g 1 b OG 1 Y g 2 a g 1 a

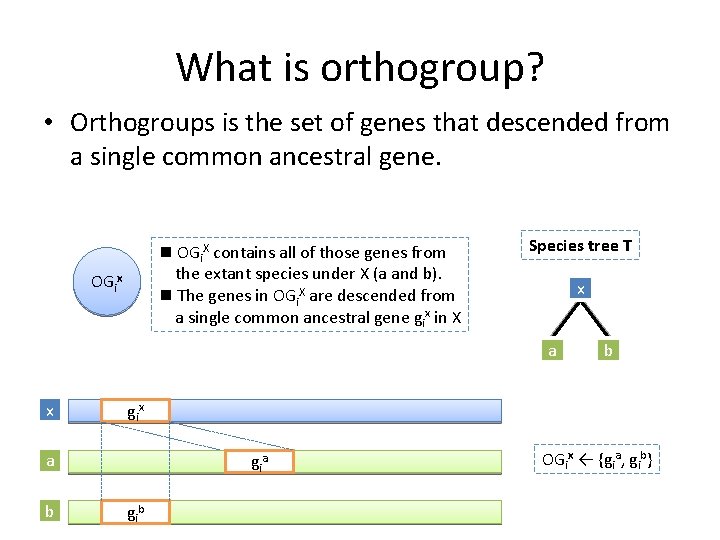
What is orthogroup? • Orthogroups is the set of genes that descended from a single common ancestral gene. n OGi. X contains all of those genes from the extant species under X (a and b). n The genes in OGi. X are descended from a single common ancestral gene gix in X OGix Species tree T x a x gi x a b b gi a gi b OGix ← {gia, gib}

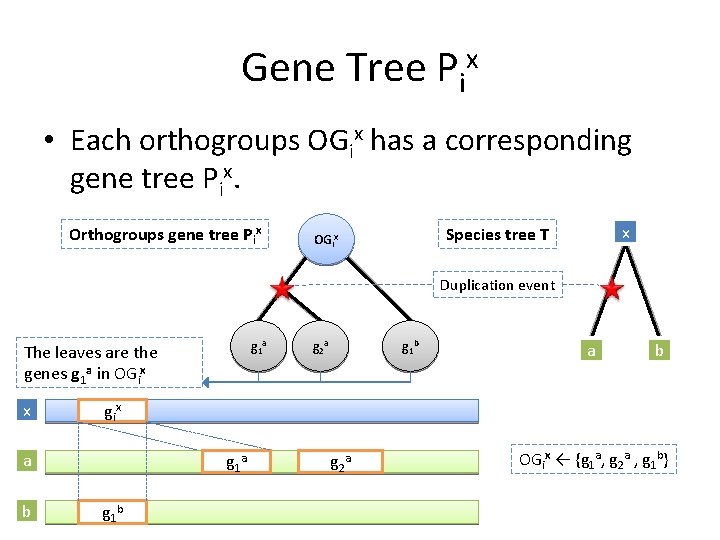
Gene Tree Pix • Each orthogroups OGix has a corresponding gene tree Pix. Orthogroups gene tree Pix x Species tree T OGix Duplication event g 1 a The leaves are the genes g 1 a in OGix x g 1 b a b gi x a b g 2 a g 1 b g 2 a OGix ← {g 1 a, g 2 a , g 1 b}

Important Definitions • Sound orthogroup (definition 1) contains only the genes that descended from a single common ancestor (specificity) • Complete orthogroup (definition 2) contains all the genes that descended from a single common ancestor (sensitivity) ※ the importance of two definitions will be discussed later

Outline • Introduction • Objective • Methods – Background Knowledge – Introduce Orthogroup – SYNERGY Algorithm • Results • Conclusion • Discussion

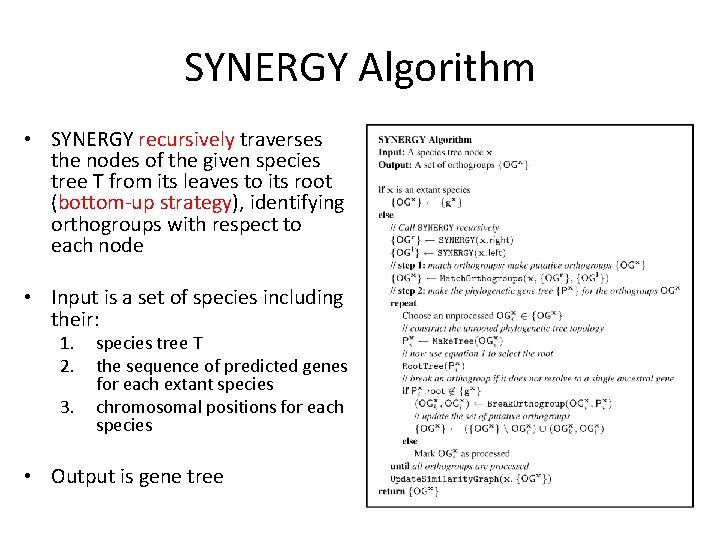
SYNERGY Algorithm • SYNERGY recursively traverses the nodes of the given species tree T from its leaves to its root (bottom-up strategy), identifying orthogroups with respect to each node • Input is a set of species including their: 1. 2. 3. species tree T the sequence of predicted genes for each extant species chromosomal positions for each species • Output is gene tree

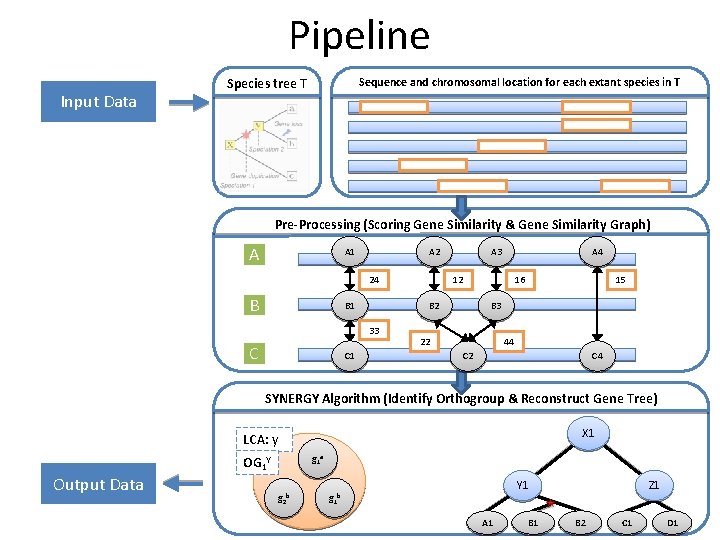
Pipeline Input Data Species tree T Sequence and chromosomal location for each extant species in T Pre-Processing (Scoring Gene Similarity & Gene Similarity Graph) A A 1 A 2 24 B B 1 C 1 A 4 12 B 2 33 C A 3 16 15 B 3 22 44 C 2 C 4 SYNERGY Algorithm (Identify Orthogroup & Reconstruct Gene Tree) X 1 LCA: y Output Data g 1 a OG 1 Y g 2 b Z 1 Y 1 g 1 b A 1 B 2 C 1 D 1

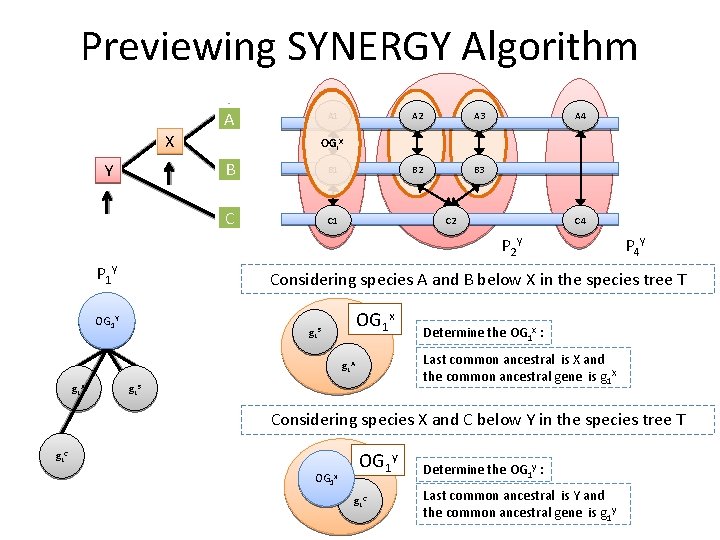
Previewing SYNERGY Algorithm A X Y A 1 A 2 A 3 B 2 B 3 A 4 OGi. X B B 1 C C 1 C 2 C 4 P 2 Y P 1 Y Considering species A and B below X in the species tree T OG 1 XY OG 1 x g 1 B Determine the OG 1 x : Last common ancestral is X and the common ancestral gene is g 1 X g 1 A P 4 Y g 1 B Considering species X and C below Y in the species tree T g 1 C OG 1 x OG 1 y g 1 C Determine the OG 1 y : Last common ancestral is Y and the common ancestral gene is g 1 y

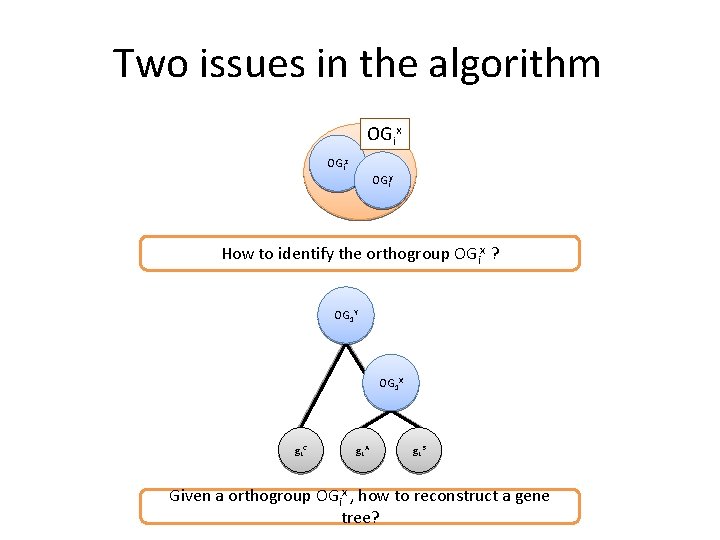
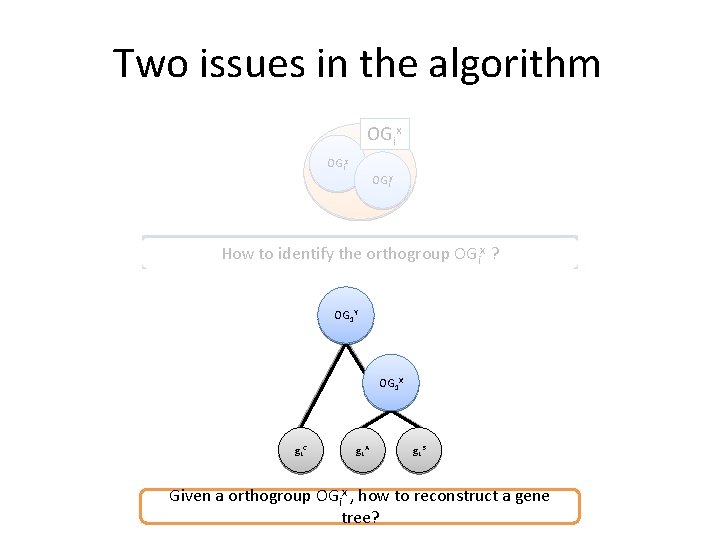
Two issues in the algorithm OGix OGiz OGiy How to identify the orthogroup OGix ? OG 1 Y OG 1 X g 1 C g 1 A g 1 B Given a orthogroup OGix , how to reconstruct a gene tree?

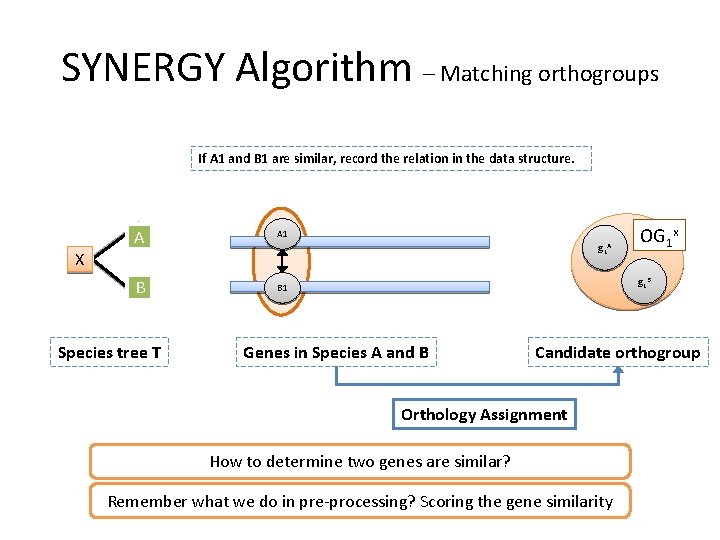
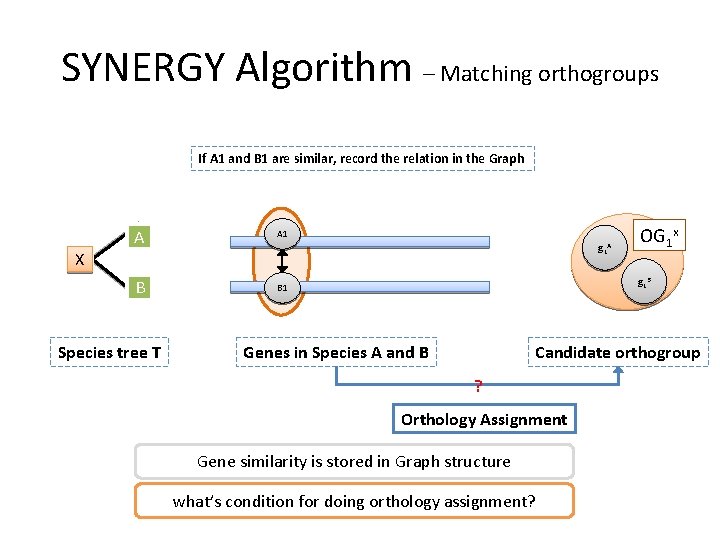
SYNERGY Algorithm – Matching orthogroups If A 1 and B 1 are similar, record the relation in the data structure. A A 1 B B 1 g 1 A X Species tree T OG 1 x g 1 B Genes in Species A and B Candidate orthogroup Orthology Assignment How to determine two genes are similar? Remember what we do in pre-processing? Scoring the gene similarity

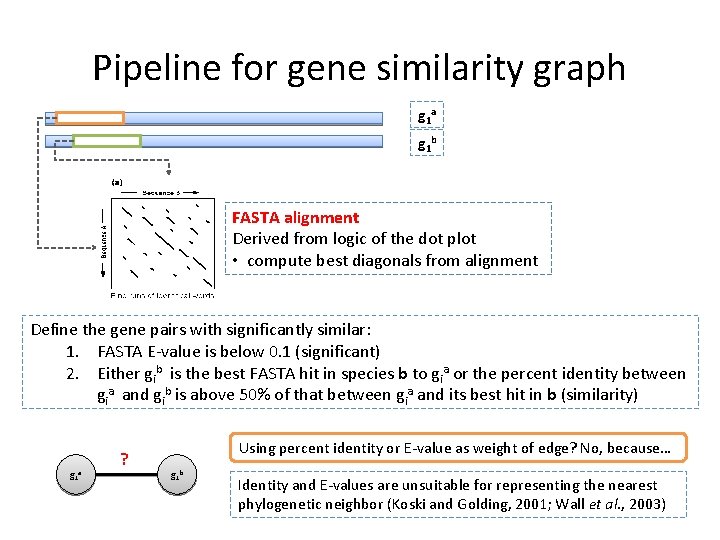
Selecting similar genes for Orthogroup • SYNERGY relies on the pre-computed distances between genes to make orthology assignments. • Execute all-versus-all FASTA alignments between all genes in the input. • The relations between genes will represented by a gene similarity graph as a weighted directed graph G = (V, E)

Pipeline for gene similarity graph g 1 a g 1 b FASTA alignment Derived from logic of the dot plot • compute best diagonals from alignment Define the gene pairs with significantly similar: 1. FASTA E-value is below 0. 1 (significant) 2. Either gib is the best FASTA hit in species b to gia or the percent identity between gia and gib is above 50% of that between gia and its best hit in b (similarity) g 1 a ? Using percent identity or E-value as weight of edge? No, because… g 1 b Identity and E-values are unsuitable for representing the nearest phylogenetic neighbor (Koski and Golding, 2001; Wall et al. , 2003)

How to get the weight of edge? • Weighting each edge by the distance similarity method. • Pre-compute peptide sequence similarity and synteny similarity scores as distance between two genes. • Peptide sequence score – globally align two proteins based on JTT amino acid substitution matrix. • Synteny similarity score – the fraction of their neighbors that are orthologous to each other

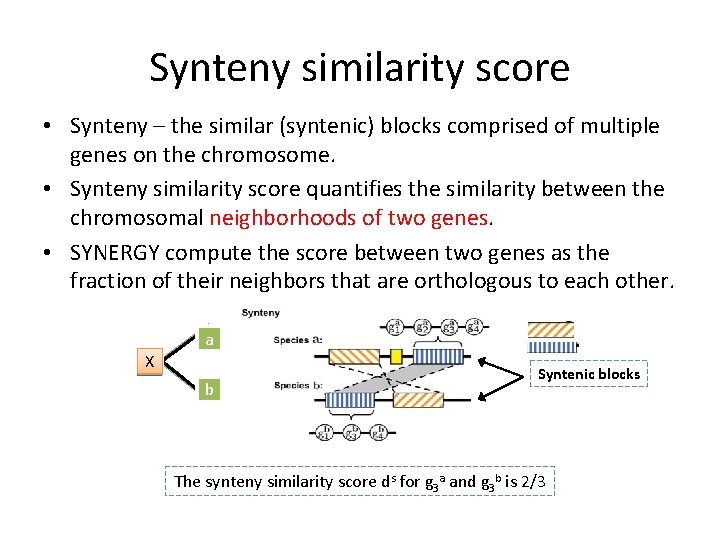
Synteny similarity score • Synteny – the similar (syntenic) blocks comprised of multiple genes on the chromosome. • Synteny similarity score quantifies the similarity between the chromosomal neighborhoods of two genes. • SYNERGY compute the score between two genes as the fraction of their neighbors that are orthologous to each other. a X b Syntenic blocks The synteny similarity score ds for g 3 a and g 3 b is 2/3

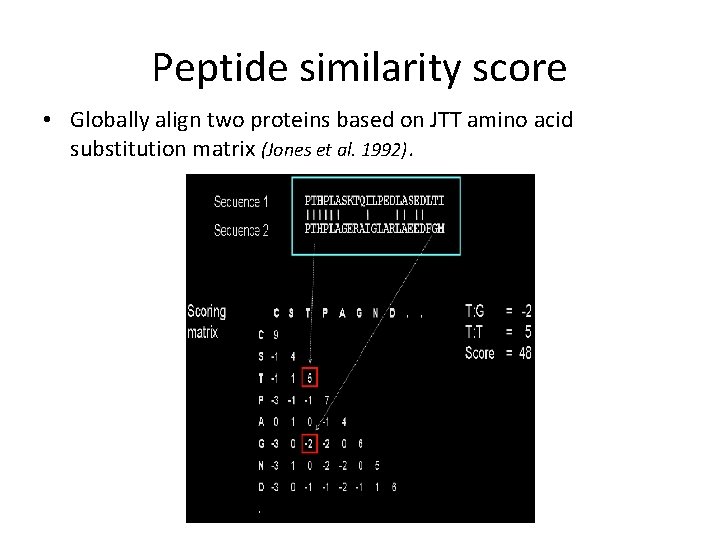
Peptide similarity score • Globally align two proteins based on JTT amino acid substitution matrix (Jones et al. 1992).

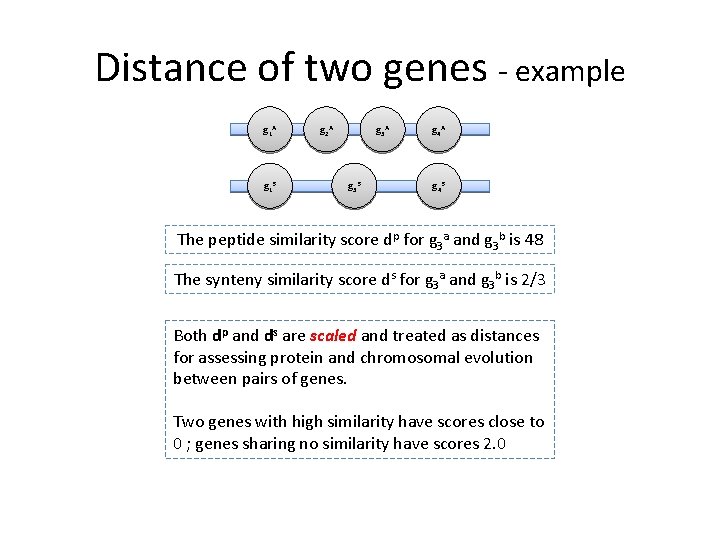
Distance of two genes - example g 1 A g 1 B g 2 A g 3 B g 4 A g 4 B The peptide similarity score dp for g 3 a and g 3 b is 48 The synteny similarity score ds for g 3 a and g 3 b is 2/3 Both dp and ds are scaled and treated as distances for assessing protein and chromosomal evolution between pairs of genes. Two genes with high similarity have scores close to 0 ; genes sharing no similarity have scores 2. 0

SYNERGY Algorithm – Matching orthogroups If A 1 and B 1 are similar, record the relation in the Graph A A 1 B B 1 g 1 A X Species tree T OG 1 x g 1 B Genes in Species A and B Candidate orthogroup ? Orthology Assignment Gene similarity is stored in Graph structure what’s condition for doing orthology assignment?

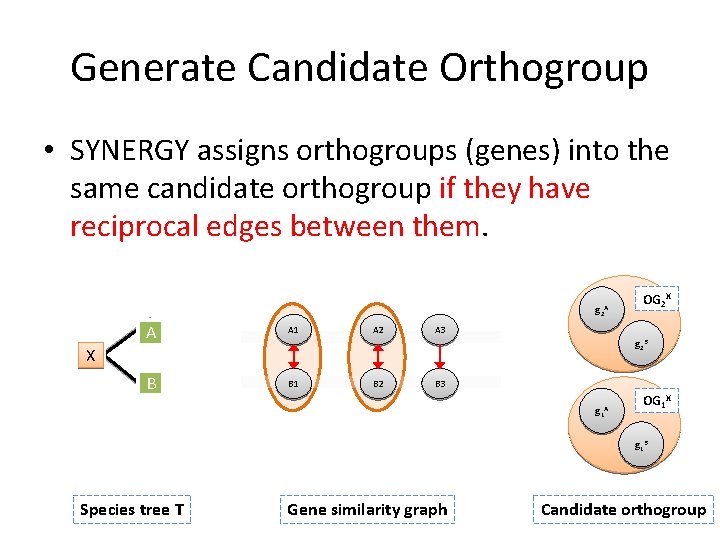
Generate Candidate Orthogroup • SYNERGY assigns orthogroups (genes) into the same candidate orthogroup if they have reciprocal edges between them. g 2 A A A 1 B B 1 A 2 OG 2 X A 3 g 2 B X B 2 B 3 g 1 A OG 1 X g 1 B Species tree T Gene similarity graph Candidate orthogroup

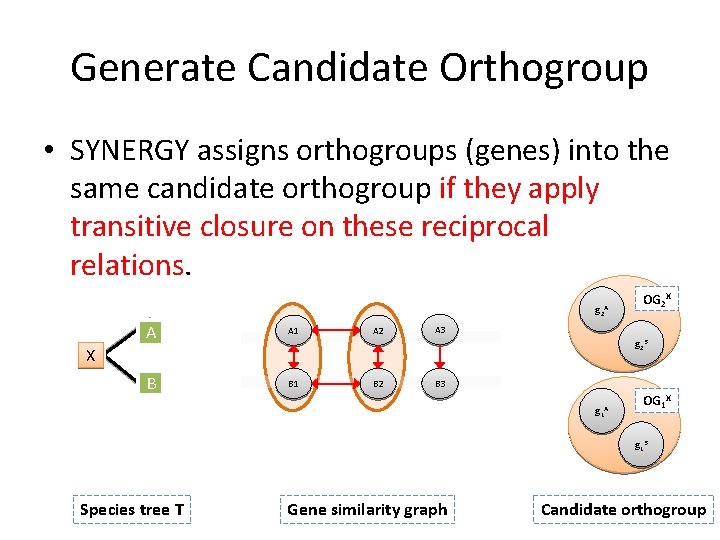
Generate Candidate Orthogroup • SYNERGY assigns orthogroups (genes) into the same candidate orthogroup if they apply transitive closure on these reciprocal relations. g 2 A A A 1 B B 1 A 2 OG 2 X A 3 g 2 B X B 2 B 3 g 1 A OG 1 X g 1 B Species tree T Gene similarity graph Candidate orthogroup

Two issues in the algorithm OGix OGiz OGiy How to identify the orthogroup OGix ? OG 1 Y OG 1 X g 1 C g 1 A g 1 B Given a orthogroup OGix , how to reconstruct a gene tree?

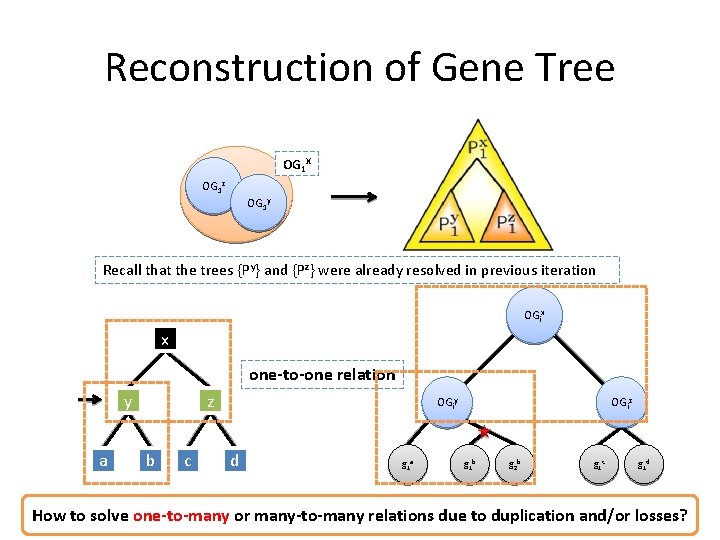
Reconstruction of Gene Tree OG 1 X OG 1 z OG 1 y Recall that the trees {Py} and {Pz} were already resolved in previous iteration OGix x one-to-one relation y a z b c OGiy d g 1 a OGiz g 1 b g 2 b g 1 c g 1 d How to solve one-to-many or many-to-many relations due to duplication and/or losses?

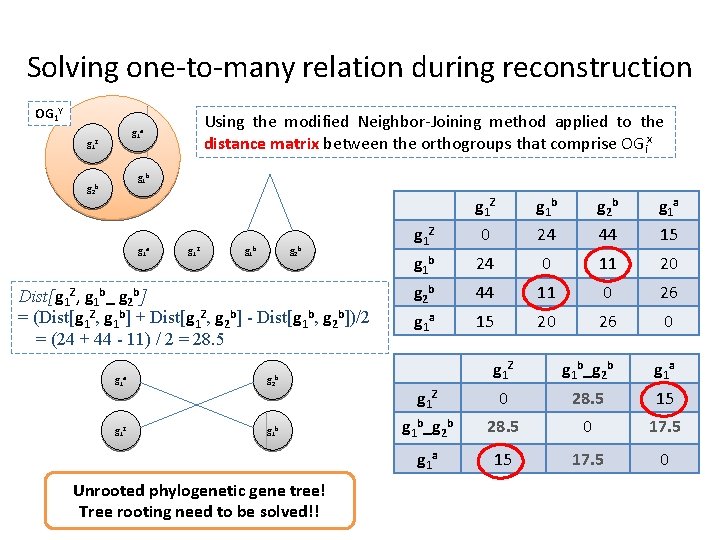
Solving one-to-many relation during reconstruction OG 1 Y Using the modified Neighbor-Joining method applied to the distance matrix between the orthogroups that comprise OGix g 1 a g 1 Z g 1 b g 2 b Dist[g 1 Z, g 1 b_ g 2 b] = (Dist[g 1 Z, g 1 b] + Dist[g 1 Z, g 2 b] - Dist[g 1 b, g 2 b])/2 = (24 + 44 - 11) / 2 = 28. 5 g 1 a g 2 b g 1 Z g 1 b Unrooted phylogenetic gene tree! Tree rooting need to be solved!! g 1 Z g 1 b g 2 b g 1 a g 1 Z 0 24 44 15 g 1 b 24 0 11 20 g 2 b 44 11 0 26 g 1 a 15 20 26 0 g 1 Z g 1 b_g 2 b g 1 a g 1 Z 0 28. 5 15 g 1 b_g 2 b 28. 5 0 17. 5 g 1 a 15 17. 5 0

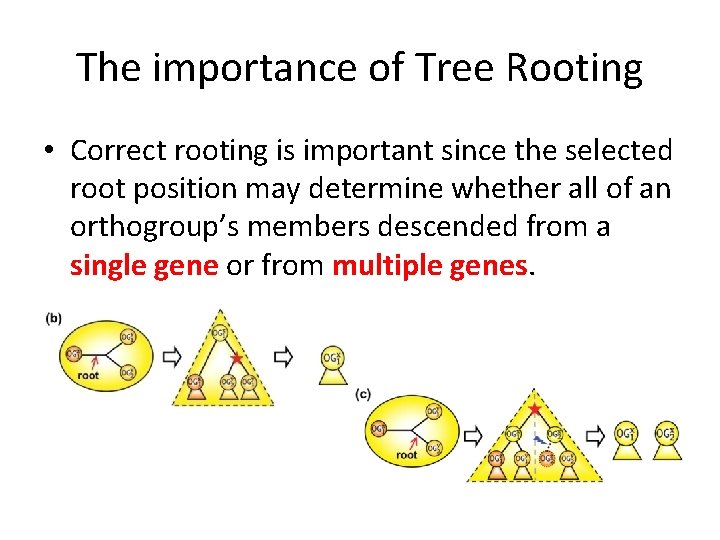
The importance of Tree Rooting • Correct rooting is important since the selected root position may determine whether all of an orthogroup’s members descended from a single gene or from multiple genes.

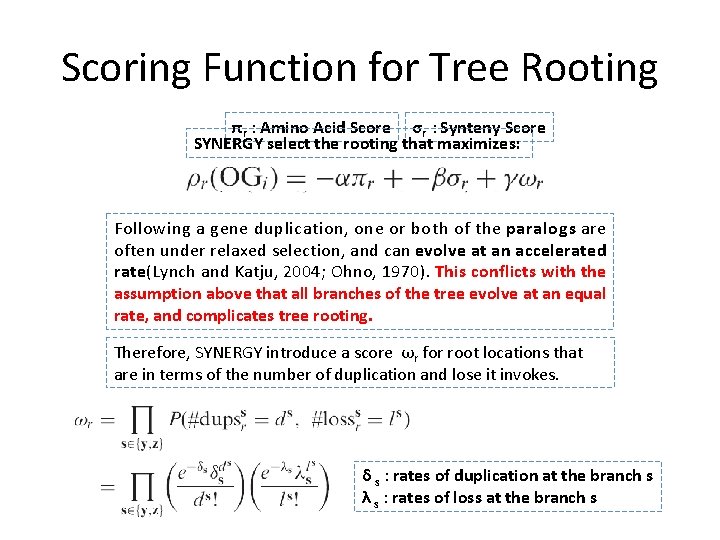
How to choose a tree’s root? • Assumption: Rates of evolution among all the leaves in a tree is equal. A tree’s root should be approximately equidistant to all the leaves. • SYNERGY compute every possible rooting r at internal branch, and assign a score to each rooting. • The score is proportional to the variance in both peptide sequence and synteny scores, termed πr and σr, respectively.

Scoring Function for Tree Rooting πr : Amino Acid Score σr : Synteny Score SYNERGY select the rooting that maximizes: Following a gene duplication, one or both of the paralogs are often under relaxed selection, and can evolve at an accelerated rate(Lynch and Katju, 2004; Ohno, 1970). This conflicts with the assumption above that all branches of the tree evolve at an equal rate, and complicates tree rooting. Therefore, SYNERGY introduce a score ωr for root locations that are in terms of the number of duplication and lose it invokes. δ s : rates of duplication at the branch s λ s : rates of loss at the branch s

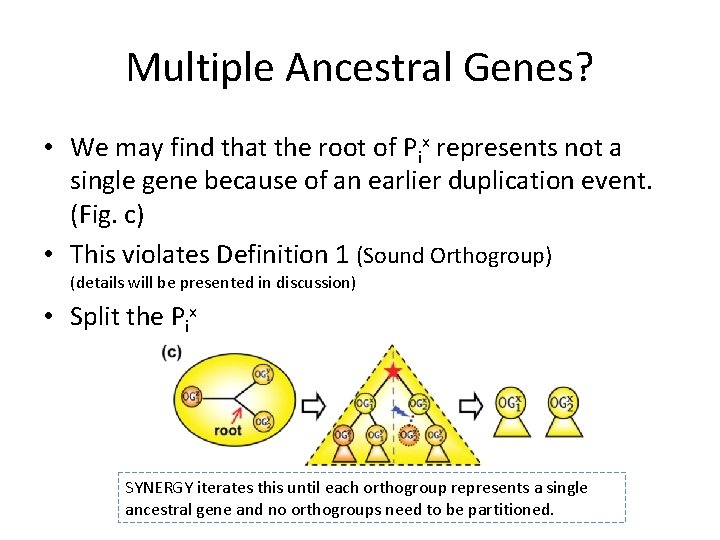
Multiple Ancestral Genes? • We may find that the root of Pix represents not a single gene because of an earlier duplication event. (Fig. c) • This violates Definition 1 (Sound Orthogroup) (details will be presented in discussion) • Split the Pix SYNERGY iterates this until each orthogroup represents a single ancestral gene and no orthogroups need to be partitioned.

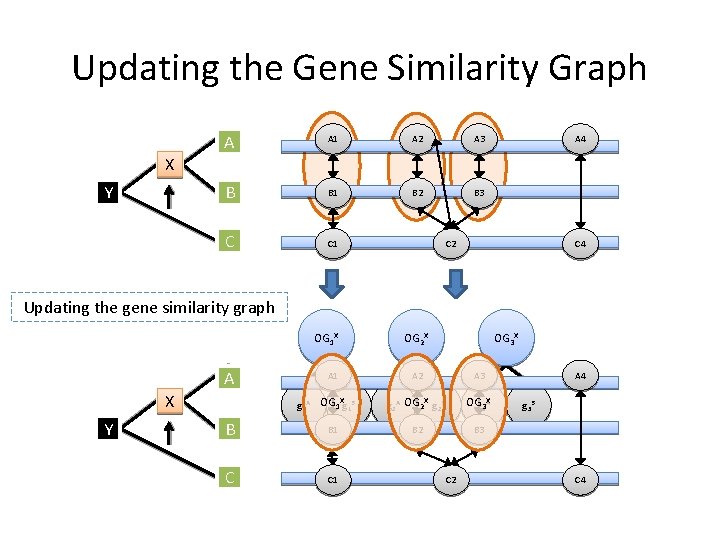
Updating the Gene Similarity Graph A A 1 A 2 A 3 B B 1 B 2 B 3 C C 1 A 4 X Y C 2 C 4 Updating the gene similarity graph OG 1 X A X Y A 1 X g 1 A OG 1 g 1 B B B 1 C C 1 OG 2 X OG 3 X A 2 A 3 X g 2 A OG 2 g 2 B OGg 33 XA B 2 B 3 C 2 A 4 g 3 B C 4

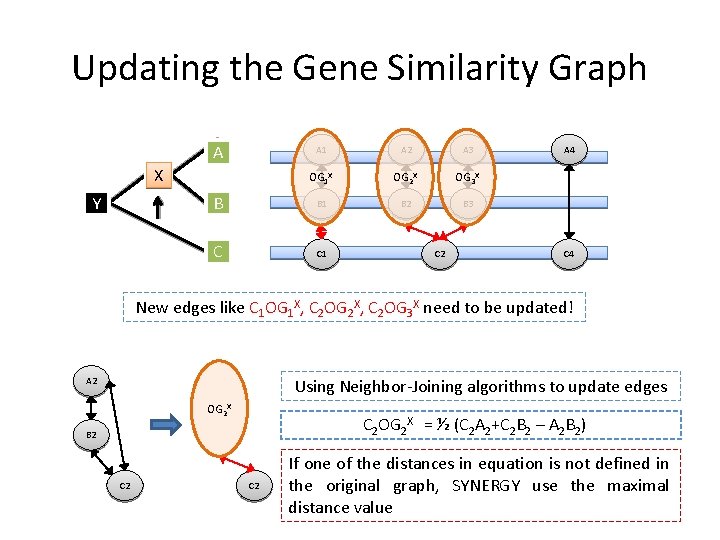
Updating the Gene Similarity Graph A A 1 A 2 A 3 OG 1 X OG 2 X OG 3 X B B 1 B 2 B 3 C C 1 X Y C 2 A 4 C 4 New edges like C 1 OG 1 X, C 2 OG 2 X, C 2 OG 3 X need to be updated! Using Neighbor-Joining algorithms to update edges A 2 OG 2 X C 2 OG 2 X = ½ (C 2 A 2+C 2 B 2 – A 2 B 2) B 2 C 2 If one of the distances in equation is not defined in the original graph, SYNERGY use the maximal distance value

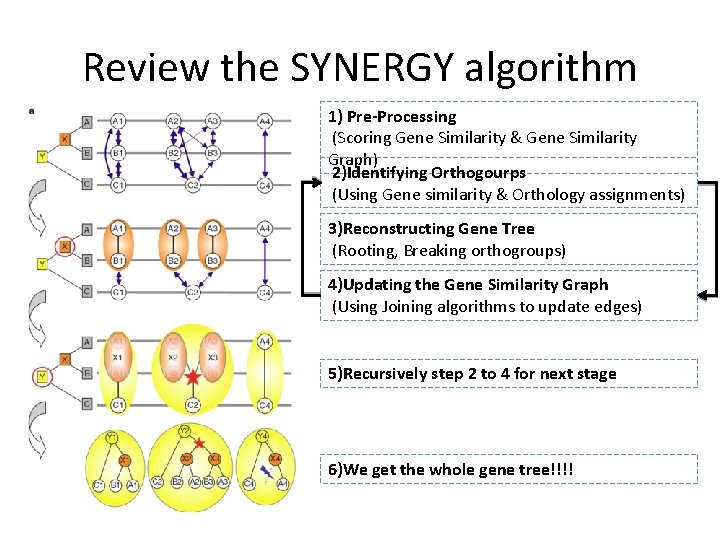
Review the SYNERGY algorithm 1) Pre-Processing (Scoring Gene Similarity & Gene Similarity Graph) 2)Identifying Orthogourps (Using Gene similarity & Orthology assignments) 3)Reconstructing Gene Tree (Rooting, Breaking orthogroups) 4)Updating the Gene Similarity Graph (Using Joining algorithms to update edges) 5)Recursively step 2 to 4 for next stage 6)We get the whole gene tree!!!!

Outline • • Introduction Objective Methods Results – Test on Ascomycota fungi – Comparison to curated resource • Conclusion • Discussion

SYNERGY Application on fungal species • Why Fungi? – With whole-genome duplication (WGD) event, followed by widespread loss of paralogous genes (Byrne and Wolfe, 2005; Dietrich et al. , 2004; Kellis et al. , 2004) – With studied model, Saccharomyces cerevisiae, offers studies of genome evolution and function (kellis et al. , 2003) • Source: nine Ascomycota fungal species with a total of 52, 092 protein coding genes

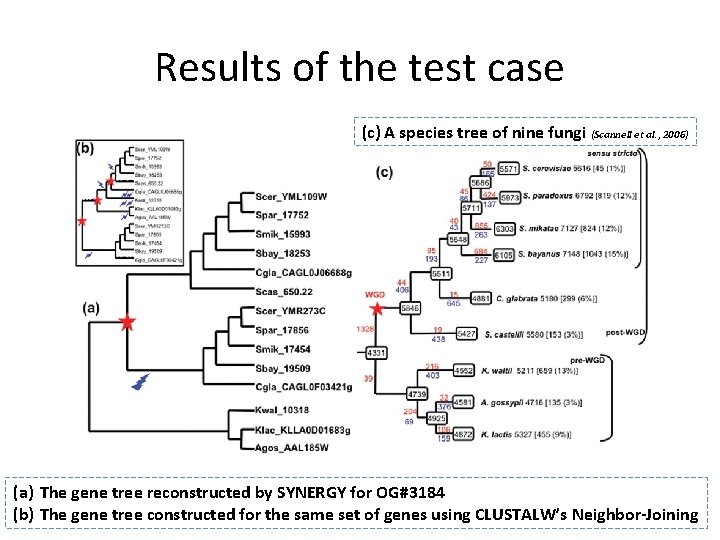
Results of the test case (c) A species tree of nine fungi (Scannell et al. , 2006) (a) The gene tree reconstructed by SYNERGY for OG#3184 (b) The gene tree constructed for the same set of genes using CLUSTALW’s Neighbor-Joining

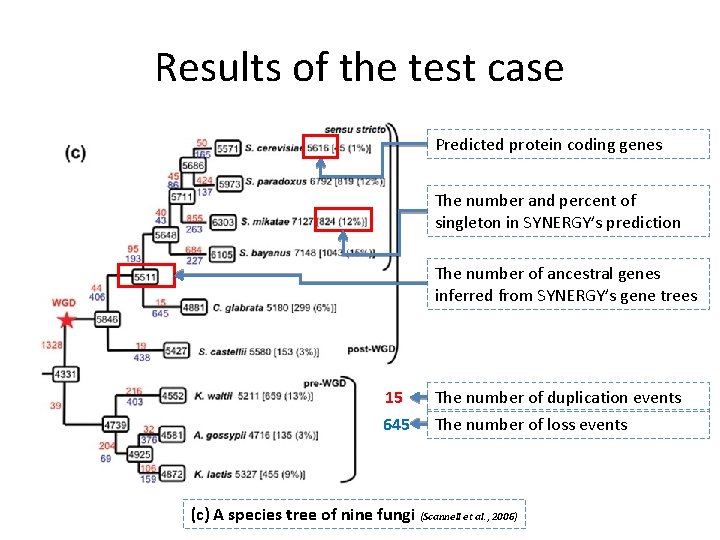
Results of the test case Predicted protein coding genes The number and percent of singleton in SYNERGY’s prediction The number of ancestral genes inferred from SYNERGY’s gene trees 15 The number of duplication events 645 The number of loss events (c) A species tree of nine fungi (Scannell et al. , 2006)

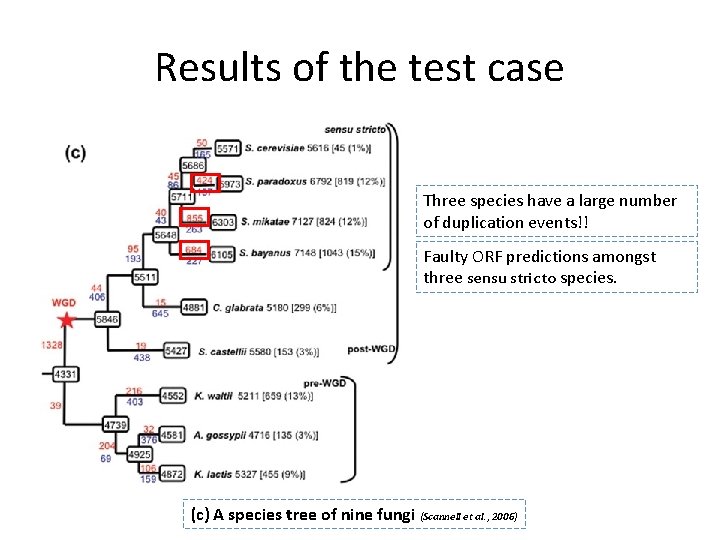
Results of the test case Three species have a large number of duplication events!! Faulty ORF predictions amongst three sensu stricto species. (c) A species tree of nine fungi (Scannell et al. , 2006)

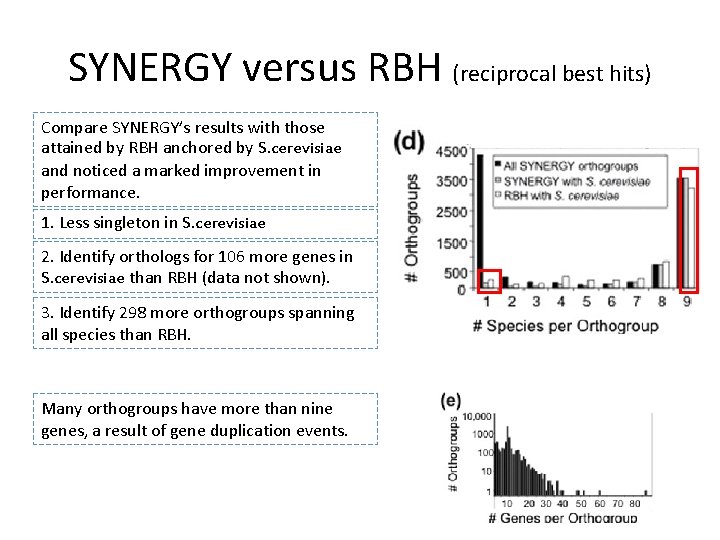
SYNERGY versus RBH (reciprocal best hits) Compare SYNERGY’s results with those attained by RBH anchored by S. cerevisiae and noticed a marked improvement in performance. 1. Less singleton in S. cerevisiae 2. Identify orthologs for 106 more genes in S. cerevisiae than RBH (data not shown). 3. Identify 298 more orthogroups spanning all species than RBH. Many orthogroups have more than nine genes, a result of gene duplication events.

Measuring Orthogroup Robustness • Jackknife-based approach – repeatedly excluding different portions of data (perturbations) to measure orthogroup robustness to – choice of species included – the accuracy of gene predictions within each species • Estimate species confidence score by systematically hiding each branch of the species tree T and running SYNERGY separately, resulting in 31 holdout experiments. • Estimate gene confidence score by randomly withholding a proportion of genes from each genome repeatedly. Set the probability of hiding each gene at 0. 1, and 50 holdout experiments

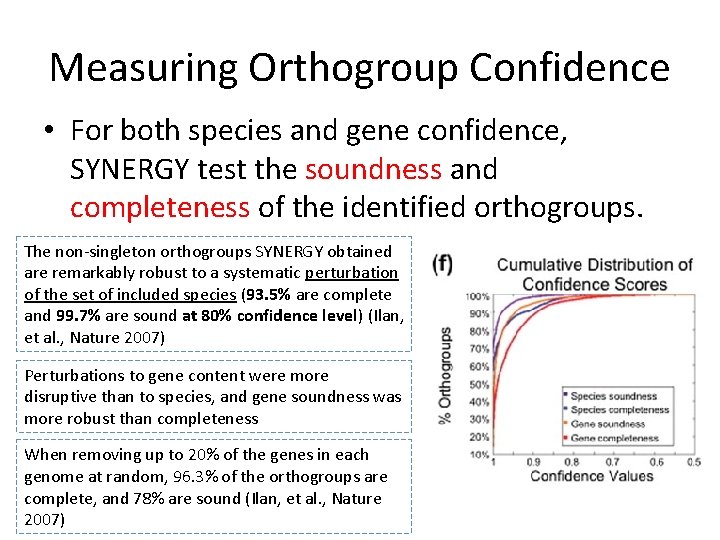
Measuring Orthogroup Confidence • For both species and gene confidence, SYNERGY test the soundness and completeness of the identified orthogroups. The non-singleton orthogroups SYNERGY obtained are remarkably robust to a systematic perturbation of the set of included species (93. 5% are complete and 99. 7% are sound at 80% confidence level) (Ilan, et al. , Nature 2007) Perturbations to gene content were more disruptive than to species, and gene soundness was more robust than completeness When removing up to 20% of the genes in each genome at random, 96. 3% of the orthogroups are complete, and 78% are sound (Ilan, et al. , Nature 2007)

Outline • • Introduction Objective Methods Results – Test on Ascomycota fungi – Comparison to curated resource • Conclusion • Discussion

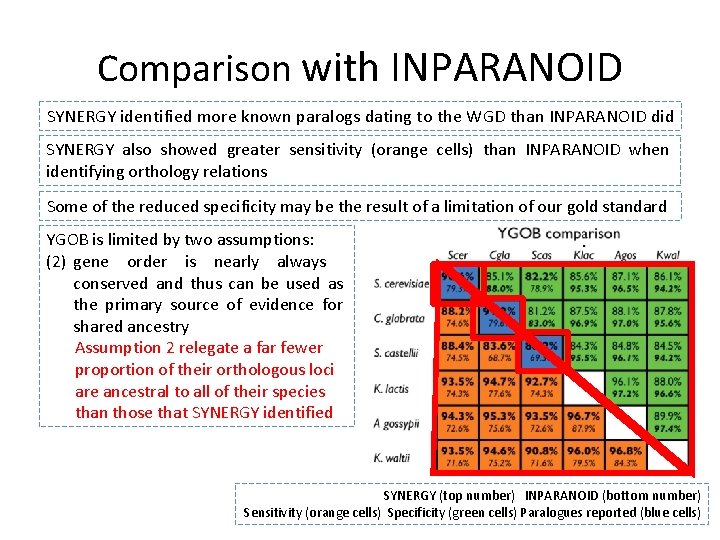
Comparison to curated resource - YGOB • Yeast Gene Order Browser (YGOB, Byrne and Wolfe, 2005), provides a “gold standard” of orthology and paralogy relations. • YGOB assume that the WGD is the only duplication event among the lineage and relies predominantly on synteny to assign orthology relations (manually curated). • Authors also compared the quality of SYNERGY’s paralogy assignments to that of INPARANOID (Remm et al. , 2001). • INPARANOID is a hit-clustering method designed to identify paralogous relations.

Comparison with INPARANOID SYNERGY identified more known paralogs dating to the WGD than INPARANOID did SYNERGY also showed greater sensitivity (orange cells) than INPARANOID when identifying orthology relations Some of the reduced specificity may be the result of a limitation of our gold standard YGOB is limited by two assumptions: (1) gene (2) all duplication order isevents nearly originated always in the WGD and conserved and thus can orthology be used is as at mostprimary the a two-to-one sourcerelationship of evidence for shared Assumption ancestry 1 relegate a greater portion Assumption of 2 genes relegateasa farsingletons fewer without proportion orthologs of their orthologous loci are ancestral to all of their species than those that SYNERGY identified SYNERGY (top number) INPARANOID (bottom number) Sensitivity (orange cells) Specificity (green cells) Paralogues reported (blue cells)

Outline • • • Introduction Objective Methods Results Conclusion Discussion

Conclusion • SYNERGY is the gnome-wide reconstruction of homology relations across multiple genomes. • SYNERGY combines hit-clustering approaches with the phylogenetic reconstruction of treebased methods. • The results of SYNERGY markedly improve over the widely used RBH approach. Also, they are comparable quality to a manually curated gold standard.

Outline • • • Introduction Objective Methods Results Conclusion Discussion

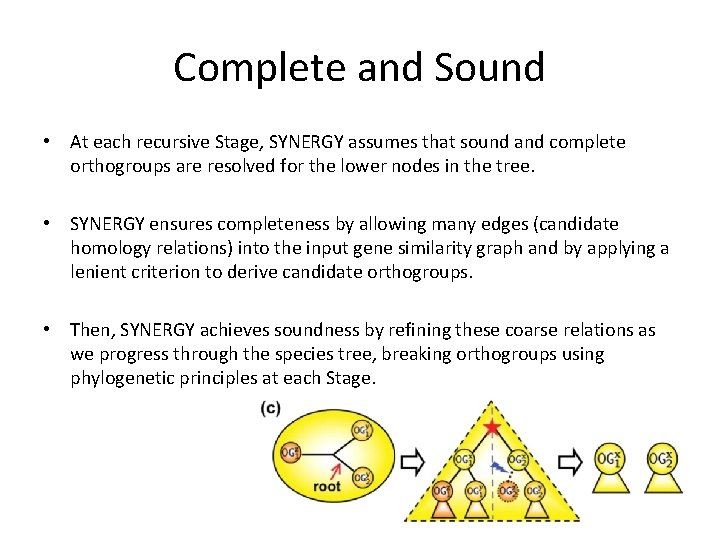
Complete and Sound • At each recursive Stage, SYNERGY assumes that sound and complete orthogroups are resolved for the lower nodes in the tree. • SYNERGY ensures completeness by allowing many edges (candidate homology relations) into the input gene similarity graph and by applying a lenient criterion to derive candidate orthogroups. • Then, SYNERGY achieves soundness by refining these coarse relations as we progress through the species tree, breaking orthogroups using phylogenetic principles at each Stage.

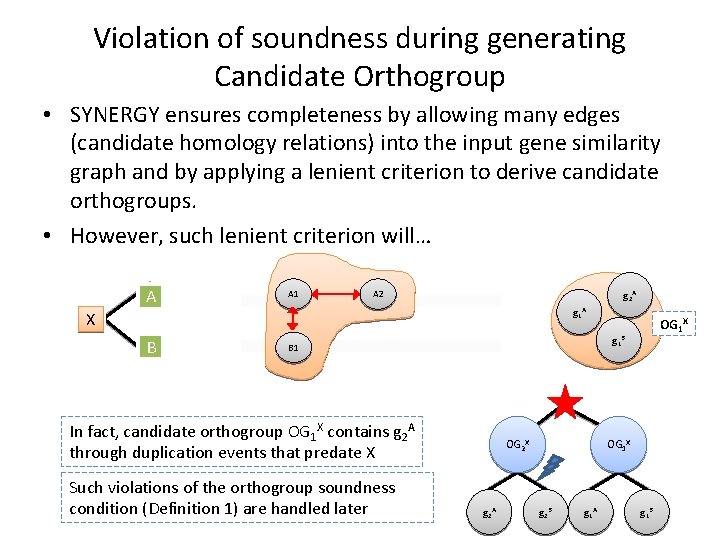
Violation of soundness during generating Candidate Orthogroup • SYNERGY ensures completeness by allowing many edges (candidate homology relations) into the input gene similarity graph and by applying a lenient criterion to derive candidate orthogroups. • However, such lenient criterion will… A A 1 A 2 g 2 A g 1 A X B g 1 B B 1 In fact, candidate orthogroup OG 1 X contains g 2 A through duplication events that predate X Such violations of the orthogroup soundness condition (Definition 1) are handled later OG 1 X OG 2 X g 2 A OG 1 X g 2 B g 1 A g 1 B

Thanks for Your Attention!

Orthogroup Confidence • A complete orthogroup (Definition 2) contains all the genes that descended from a single common ancestor and thus its genes should not ‘migrate out’ of it in the holdout experiments. • where, h(gj , gk) and OGi (gj , gk) specify the last species in the tree in which gj and gk share a common ancestor in the holdout experiment h and the original orthogroup

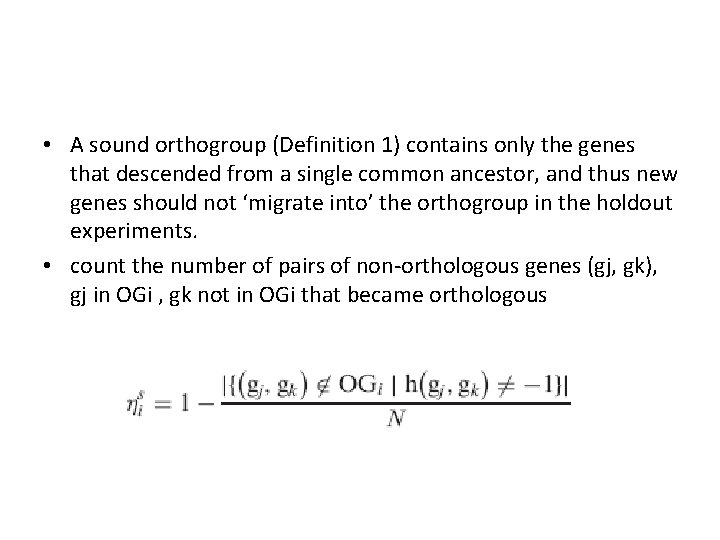
• A sound orthogroup (Definition 1) contains only the genes that descended from a single common ancestor, and thus new genes should not ‘migrate into’ the orthogroup in the holdout experiments. • count the number of pairs of non-orthologous genes (gj, gk), gj in OGi , gk not in OGi that became orthologous

Introduction • Current methods for inferring homology relation – Pair-wise sequence comparison • Best bi-directional BLAST hits • Focuses on one-to-one orthologs (no duplications) – Hit clustering methods • Detect clusters in graph of pair-wise hits – Synteny methods • Detect conserved regions, stretches of nearby hits – Phylogenetic methods • Phylogeny of family clusters orthologs near each other