Automated Annotation of Microbial Genomes Opportunities and Pitfalls

Automated Annotation of Microbial Genomes, Opportunities and Pitfalls Margie Romine Pacific Northwest National Laboratory Richland, Washington
Shewanella oneidensis MR-1 • Breathes Mn & Fe and other metals thereby changing their solubility • Also reduces radionuclides and hence impacts their mobility at contaminated sites • Genome sequenced by the Institute for Genome Research in 2002 (funded by DOE-OBER) • Can we now better determine how this organism interacts with metals and radionuclides?
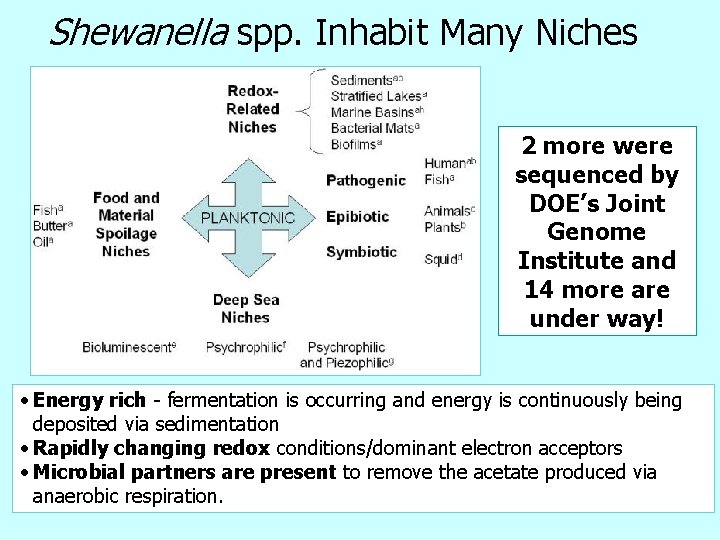
Shewanella spp. Inhabit Many Niches 2 more were sequenced by DOE’s Joint Genome Institute and 14 more are under way! • Energy rich - fermentation is occurring and energy is continuously being deposited via sedimentation • Rapidly changing redox conditions/dominant electron acceptors • Microbial partners are present to remove the acetate produced via anaerobic respiration.
Bacterial Genome Sequencing Explodes • 341 completed genomes, 976 ongoing • Partial genome sequences released in just days now by JGI! • How do we use sequence information to understand how all these organisms function in the environment? • Annotation is the key, but is now largely automated and hence of lower quality
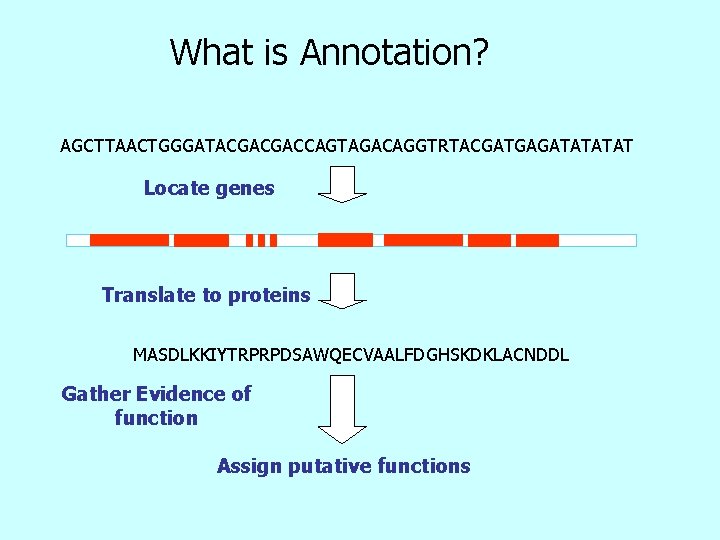
What is Annotation? AGCTTAACTGGGATACGACGACCAGTAGACAGGTRTACGATGAGATAT Locate genes Translate to proteins MASDLKKIYTRPRPDSAWQECVAALFDGHSKDKLACNDDL Gather Evidence of function Assign putative functions
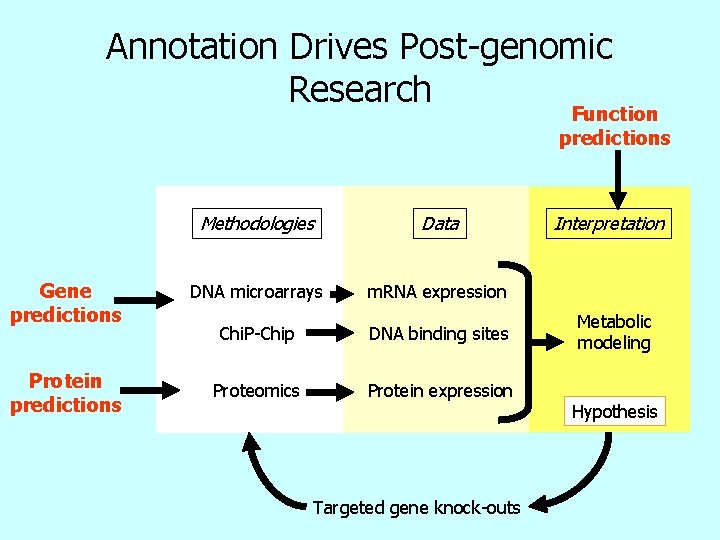
Annotation Drives Post-genomic Research Function predictions Gene predictions Protein predictions Methodologies Data DNA microarrays m. RNA expression Chi. P-Chip DNA binding sites Proteomics Protein expression Targeted gene knock-outs Interpretation Metabolic modeling Hypothesis
Annotation with Gnare/Puma 2 • Developed at Argonne National Laboratory by Natalia Maltsev, Mark D’Souza, Elizabeth Glass, Dina Sulakhe, Mustafa Syed, Pavan Anumula • http: //compbio. mcs. anl. gov/puma 2/cgibin/index. cgi • Gnare – Private genome sequences • Puma 2 – Public genome sequences
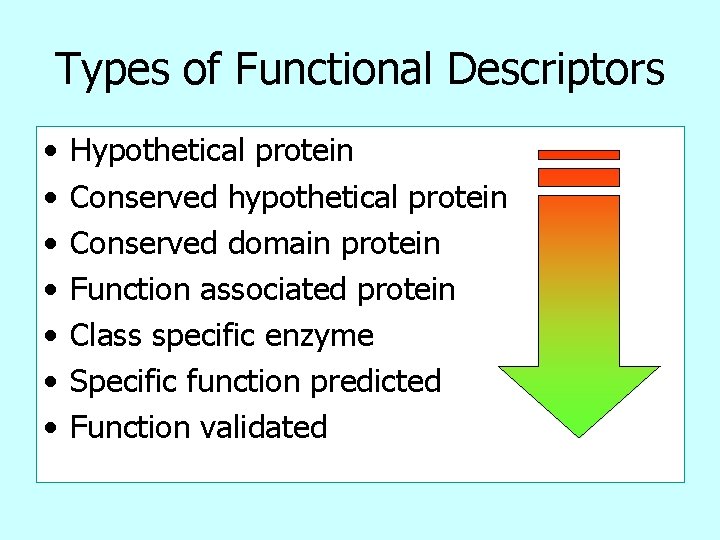
Types of Functional Descriptors • • Hypothetical protein Conserved hypothetical protein Conserved domain protein Function associated protein Class specific enzyme Specific function predicted Function validated
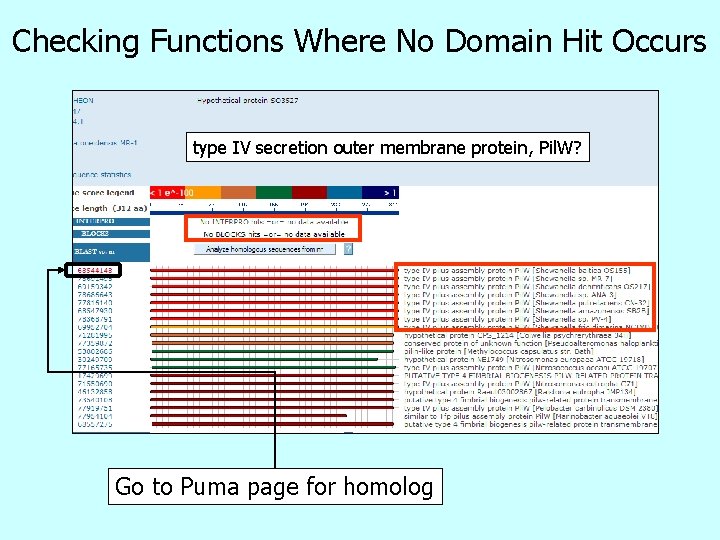
Checking Functions Where No Domain Hit Occurs type IV secretion outer membrane protein, Pil. W? Go to Puma page for homolog
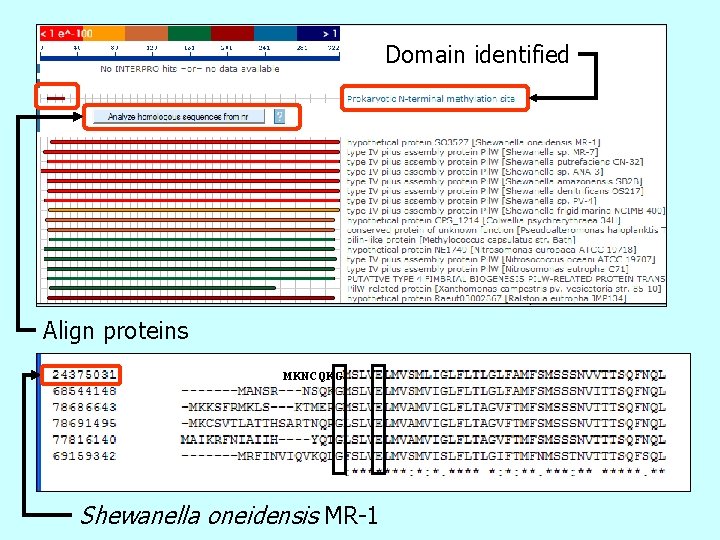
Domain identified Align proteins MKNCQKG Shewanella oneidensis MR-1
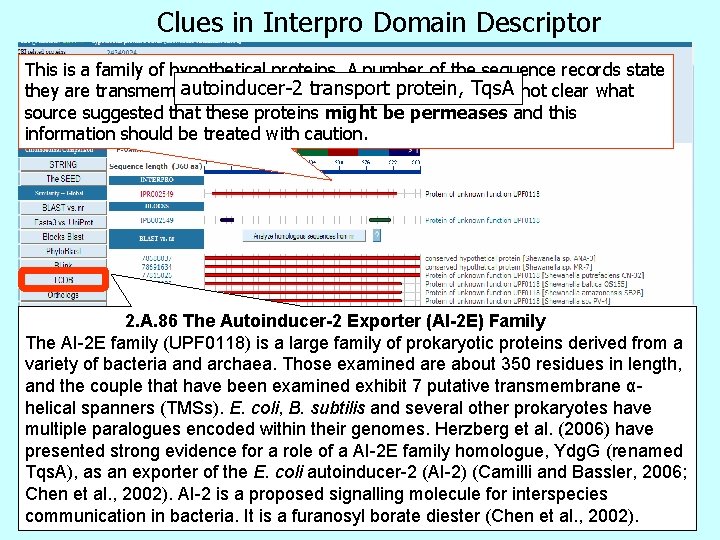
Clues in Interpro Domain Descriptor This is a family of hypothetical proteins. A number of the sequence records state autoinducer-2 protein, Tqs. A they are transmembrane proteins ortransport putative permeases. It is not clear what source suggested that these proteins might be permeases and this information should be treated with caution. 2. A. 86 The Autoinducer-2 Exporter (AI-2 E) Family The AI-2 E family (UPF 0118) is a large family of prokaryotic proteins derived from a variety of bacteria and archaea. Those examined are about 350 residues in length, and the couple that have been examined exhibit 7 putative transmembrane αhelical spanners (TMSs). E. coli, B. subtilis and several other prokaryotes have multiple paralogues encoded within their genomes. Herzberg et al. (2006) have presented strong evidence for a role of a AI-2 E family homologue, Ydg. G (renamed Tqs. A), as an exporter of the E. coli autoinducer-2 (AI-2) (Camilli and Bassler, 2006; Chen et al. , 2002). AI-2 is a proposed signalling molecule for interspecies communication in bacteria. It is a furanosyl borate diester (Chen et al. , 2002).
No functional clues Using Genome Context to Predict Function
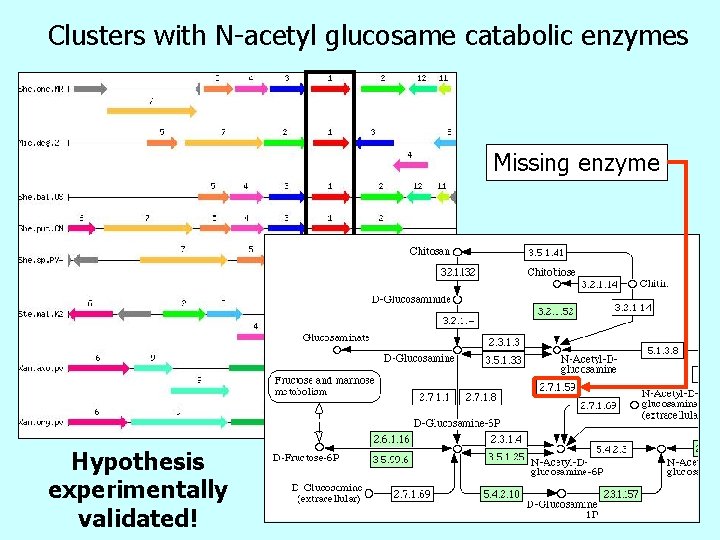
Clusters with N-acetyl glucosame catabolic enzymes Missing enzyme Hypothesis experimentally validated!
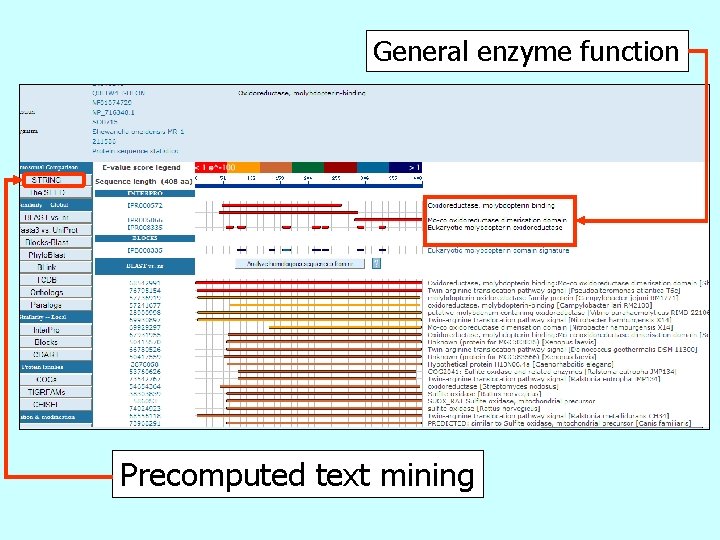
General enzyme function Precomputed text mining
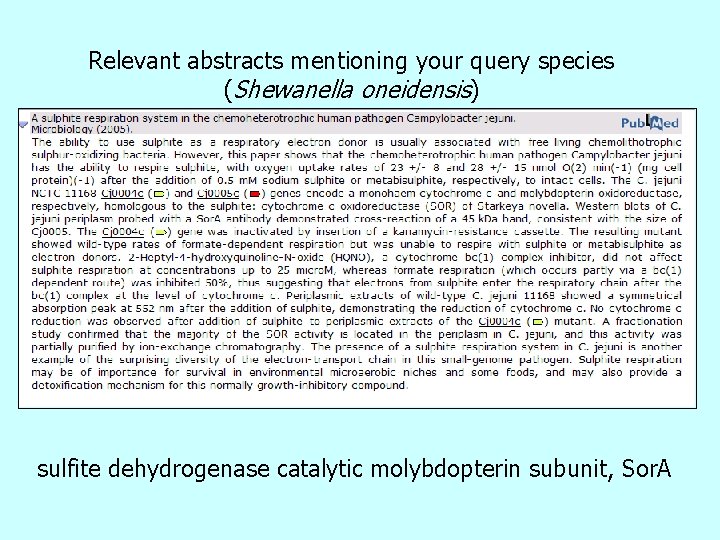
Relevant abstracts mentioning your query species (Shewanella oneidensis) sulfite dehydrogenase catalytic molybdopterin subunit, Sor. A
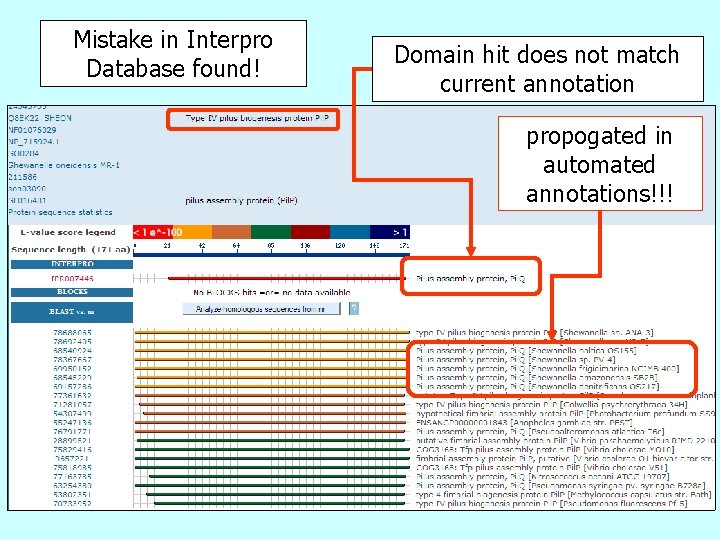
Mistake in Interpro Database found! Domain hit does not match current annotation propogated in automated annotations!!!

More Automation in Evidence Collecting Needed
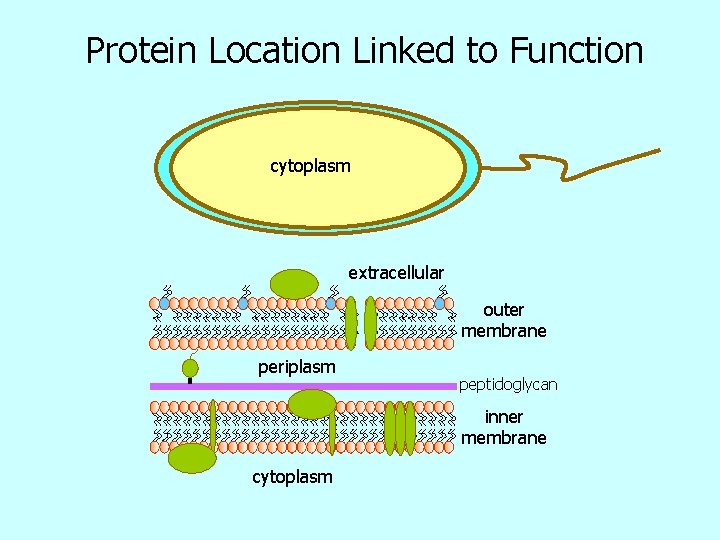
Protein Location Linked to Function cytoplasm extracellular outer membrane periplasm peptidoglycan inner membrane cytoplasm
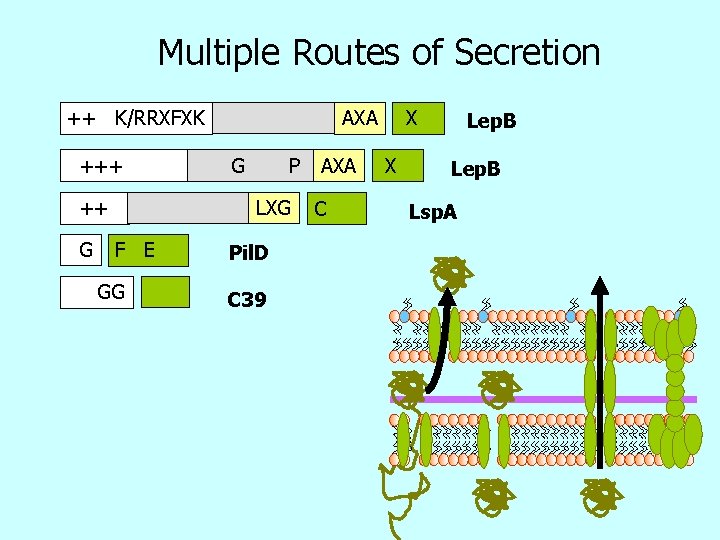
Multiple Routes of Secretion ++ K/RRXFXK +++ ++ G AXA G P LXG F E GG Pil. D C 39 AXA C X X Lep. B Lsp. A
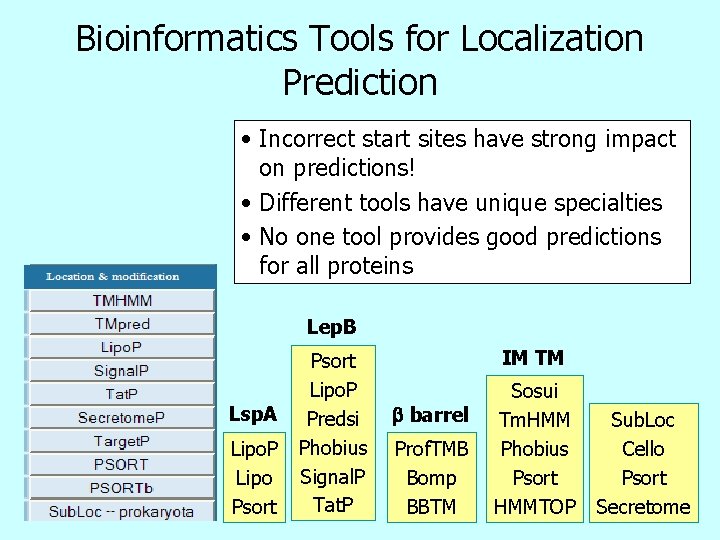
Bioinformatics Tools for Localization Prediction • Incorrect start sites have strong impact on predictions! • Different tools have unique specialties • No one tool provides good predictions for all proteins Lep. B Lsp. A Lipo. P Lipo Psort Lipo. P Predsi Phobius Signal. P Tat. P IM TM b barrel Prof. TMB Bomp BBTM Sosui Tm. HMM Phobius Psort HMMTOP Sub. Loc Cello Psort Secretome
Example: c type cytochromes • Contain CXXCH motif for binding heme …so do some other proteins that are not c type cytochromes • All are secreted across the inner membrane and then assembled • 60 proteins in MR-1 have CXXCH • Only 43 have a leader peptide and are predicted to be c type cytochromes
Future Needs in Annotation Automation • Current methods of automated annotation will lead to propagation of annotation errors and burying of useful evidence • But manual annotation cannot keep up with rate at which sequences are produced • Additional automations are needed! – Protein localization – Specialty database mining (TCDB, merops, etc) – Experimental data mining – appropriate databases don’t exist
- Slides: 22