Authentication of Human Cell Lines by DNA STR

Authentication of Human Cell Lines by DNA STR Profiling Nicole Sanderlin Field Support Scientist

STR (Short Tandem Repeat) Genotyping The LOCUS identifies the specific physical location of a gene or STR on a chromosome. The ALLELE for the gene or STR contained at that locus may be the same on both chromosomes (homozygous), or different on each chromosome (heterozygous) Proprietary Information. Not for further distribution.
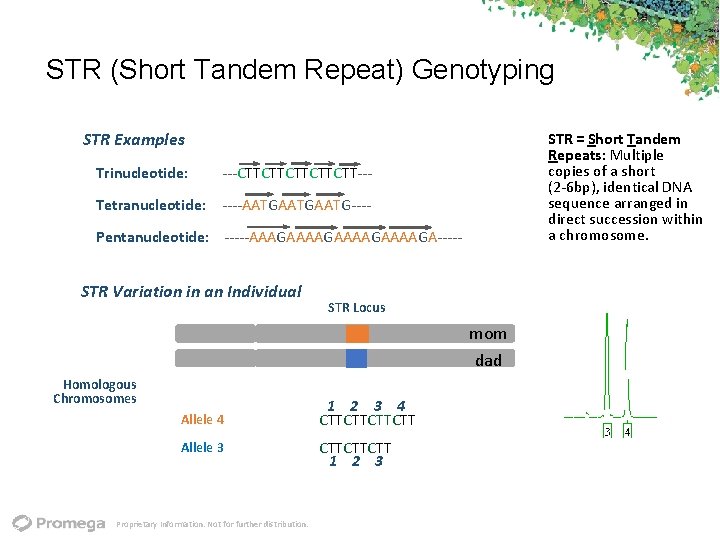
STR (Short Tandem Repeat) Genotyping STR Examples Trinucleotide: ‐‐‐CTTCTTCTT‐‐‐ Tetranucleotide: ‐‐‐‐AATGAATG‐‐‐‐ STR = Short Tandem Repeats: Multiple copies of a short (2‐ 6 bp), identical DNA sequence arranged in direct succession within a chromosome. Pentanucleotide: ‐‐‐‐‐AAAGAAAAGA‐‐‐‐‐ STR Variation in an Individual STR Locus mom dad Homologous Chromosomes Allele 4 1 2 3 4 CTTCTT Allele 3 CTTCTTCTT 1 2 3 Proprietary Information. Not for further distribution.
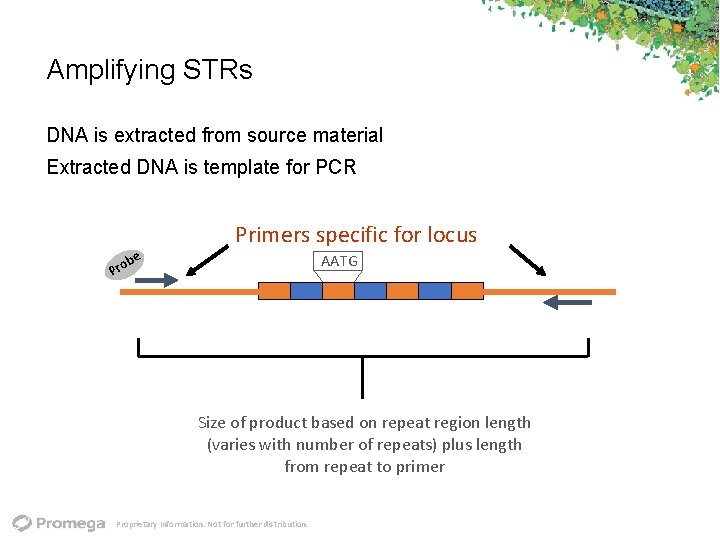
Amplifying STRs DNA is extracted from source material Extracted DNA is template for PCR Primers specific for locus e AATG b Pro Size of product based on repeat region length (varies with number of repeats) plus length from repeat to primer Proprietary Information. Not for further distribution.
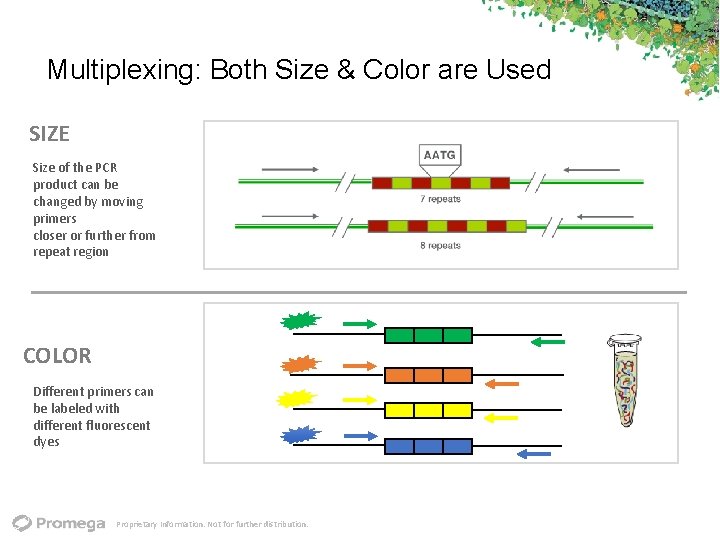
Multiplexing: Both Size & Color are Used SIZE Size of the PCR product can be changed by moving primers closer or further from repeat region COLOR Different primers can be labeled with different fluorescent dyes Proprietary Information. Not for further distribution.
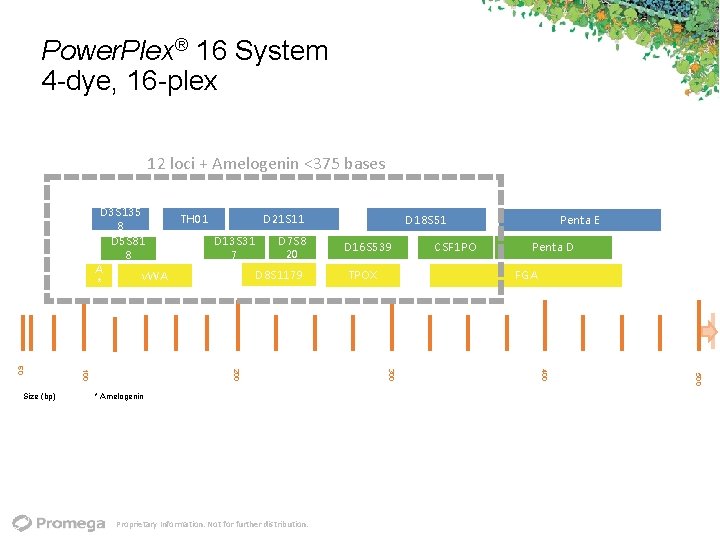
Power. Plex® 16 System 4 dye, 16 plex 12 loci + Amelogenin <375 bases D 3 S 135 8 D 5 S 81 8 A v. WA * D 21 S 11 D 13 S 31 7 D 7 S 8 20 D 8 S 1179 TPOX CSF 1 PO Penta D FGA 500 Proprietary Information. Not for further distribution. D 16 S 539 Penta E 400 * Amelogenin D 18 S 51 300 200 100 60 Size (bp) TH 01
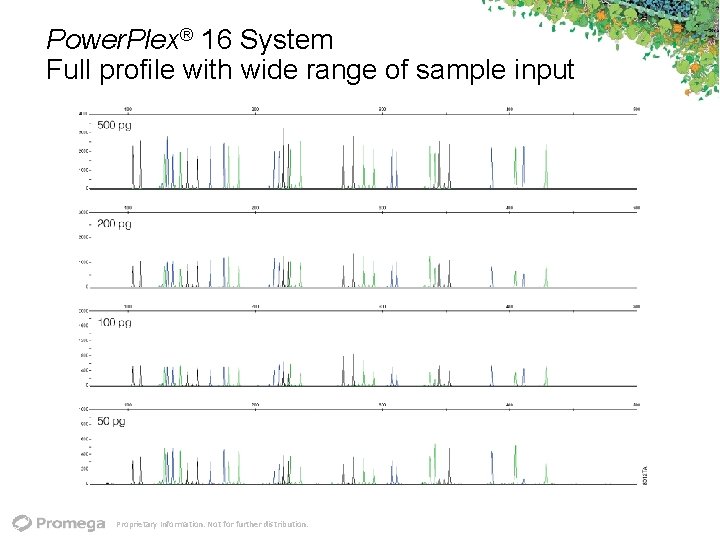
Power. Plex® 16 System Full profile with wide range of sample input Proprietary Information. Not for further distribution.
Why use STRs? • Highly informative genetic markers that are abundant and well spaced throughout the genome. • Inherited in a Mendelian manner, which makes them suitable for genetics studies. • A large number of STRs are well characterized, and allow for a high degree of multiplexing, enhancing efficiency and sample throughput. • STR genotyping is a widely accepted tool for a variety of applications such as forensic casework, paternity analysis, cell line authentication, identification of mismatch repair deficiency through microsatellite instability, linkage mapping studies and population genetics studies. Proprietary Information. Not for further distribution.

STR Profiling: Authentication Standard for Human Cell Lines

The legend of He. La • He. La first immortal cancer line to be established in 1951 by George Gey (Scherer wf, Syverton jt, Gey go. J Exp Med. 1953 97(5): 695 710. ) • Cervical cancer derived from patient Henrietta Lacks • Widely used in research. According to Rebecca Skloot “More than 60, 000 scientific articles had been published about research done on He. La, and that number was increasing steadily at a rate of more than 300 papers each month. ” • Rapid growing; can contaminate and overtake other cell types He. La cells Proprietary Information. Not for further distribution.
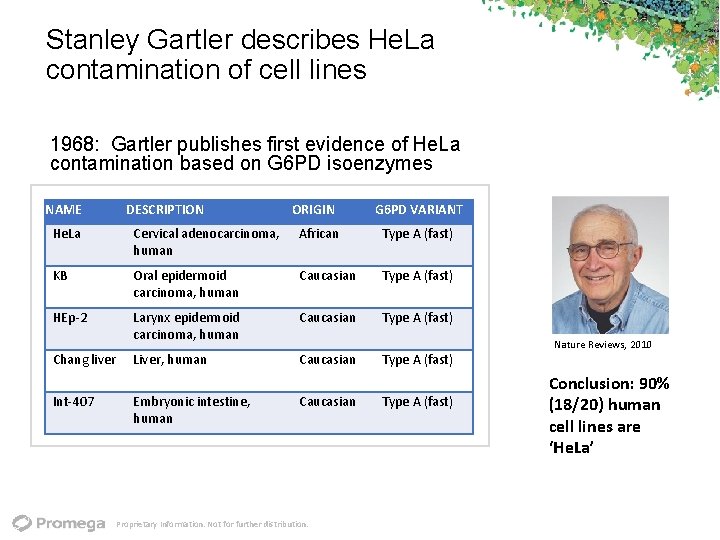
Stanley Gartler describes He. La contamination of cell lines 1968: Gartler publishes first evidence of He. La contamination based on G 6 PD isoenzymes NAME DESCRIPTION ORIGIN G 6 PD VARIANT He. La Cervical adenocarcinoma, human African Type A (fast) KB Oral epidermoid carcinoma, human Caucasian Type A (fast) HEp‐ 2 Larynx epidermoid carcinoma, human Caucasian Type A (fast) Liver, human Caucasian Chang liver Int‐ 407 Embryonic intestine, human Caucasian Proprietary Information. Not for further distribution. Type A (fast) Nature Reviews, 2010 Conclusion: 90% (18/20) human cell lines are ‘He. La’
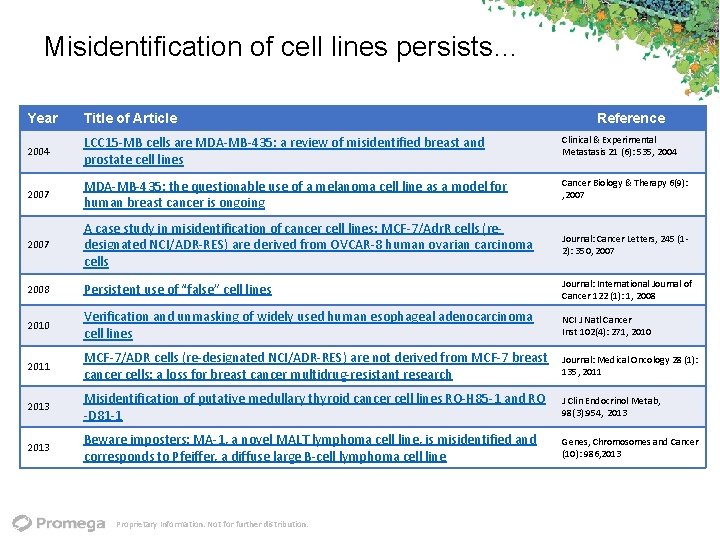
Misidentification of cell lines persists… Year Title of Article 2004 LCC 15‐MB cells are MDA‐MB‐ 435: a review of misidentified breast and prostate cell lines Clinical & Experimental Metastasis 21 (6): 535, 2004 2007 MDA‐MB‐ 435: the questionable use of a melanoma cell line as a model for human breast cancer is ongoing Cancer Biology & Therapy 6(9): , 2007 A case study in misidentification of cancer cell lines: MCF‐ 7/Adr. R cells (re‐ designated NCI/ADR‐RES) are derived from OVCAR‐ 8 human ovarian carcinoma cells Journal: Cancer Letters, 245 (1‐ 2): 350, 2007 2008 Persistent use of “false” cell lines Journal: International Journal of Cancer 122 (1): 1, 2008 2010 Verification and unmasking of widely used human esophageal adenocarcinoma cell lines NCI J Natl Cancer Inst 102(4): 271, 2010 2011 MCF‐ 7/ADR cells (re‐designated NCI/ADR‐RES) are not derived from MCF‐ 7 breast cancer cells: a loss for breast cancer multidrug‐resistant research Journal: Medical Oncology 28 (1): 135, 2011 2013 Misidentification of putative medullary thyroid cancer cell lines RO‐H 85‐ 1 and RO ‐D 81‐ 1 J Clin Endocrinol Metab, 98(3): 954, 2013 Beware imposters: MA‐ 1, a novel MALT lymphoma cell line, is misidentified and corresponds to Pfeiffer, a diffuse large B‐cell lymphoma cell line Genes, Chromosomes and Cancer (10): 986, 2013 Proprietary Information. Not for further distribution. Reference
How does false cell line identity occur? • Cross contamination during original cell line development • Cross contamination in the lab from which you received the cell line • Cross contamination in your lab • Mislabeling Proprietary Information. Not for further distribution.
Authentication Pitfalls Two cell lines with the same name • 15 C 6 (ATCC CRL 2431) mouse myeloma • 15 C 6 (ATCC HB 326) rat/mouse fusion Morphing cell line name: LLC‐MK 2 LLCMK 2 1/3 OF RESEARCHES obtain their cell lines from another lab Sarntivijai et al. Bioinformatics. 24: 2760‐ 2766, 2008 Proprietary Information. Not for further distribution.
Consequences of using misidentified cell lines • Loss of time and money • Misinformation in the public domain • Discordant or irreproducible results • Tarnished reputation © DAN PAGE COLLECTION/THEISPOT “If we’re not using what we think we are using, we’re not testing our hypotheses. We’re just gumming up the literature. I’m not sure what we’re doing, but that’s not science. ” Jeffrey Boatright, Emory Uni versity, The Big Clean Up. The Scientist Magazine. September 2015 Proprietary Information. Not for further distribution.
Impact of misidentified cell lines on applied research Experimental results based on contaminated cell lines: • Clinical trial recruiting EAC patients • 100 scientific publications • At least 3 NIH cancer research grants • 11 US patents Proprietary Information. Not for further distribution.

How researchers can prevent the use of misidentified cell lines? Ø Become aware of the magnitude of the problem Ø Authenticate all cell lines you are working with! Ø Eliminate laboratory practices that result in misidentification Ø Notify recipients of misidentified cell lines Ø Publish retractions/explanations of “false” publications Ø RESULTS MATTER Ø Confidence in the published data ONLY 10% OF AUTHORS authenticate their cell lines Ø Meet the requirements of peer reviewed journals and grant submission Ø Aid in troubleshooting undetermined or unexpected results Proprietary Information. Not for further distribution.

Nature – Policy changes From May 1 (2015), all authors of papers involving cell lines that are submitted to Nature journals will be asked whether they have checked their cell lines against publicly available lists of those known to be problematic. Proprietary Information. Not for further distribution.

Which Journals ask for Cell Line Authentication? * • AACR journals Cancer Discovery Cancer Research Clinical Cancer Research Cancer Epidemiology, Biomarkers & Prevention Molecular Cancer Research Molecular Cancer Therapeutics Cancer Prevention Research • Carcinogenesis • Cell Biochemistry and Biophysics • Cell Biology International • Endocrine Society journals Endocrinology Endocrine Reviews Journal of Clinical Endocrinology & Metabolism Molecular Endocrinology Hormones and Cancer • International Journal of Cancer • In Vitro Cellular & Developmental Biology Animal Proprietary Information. Not for further distribution. • • Journal of Molecular Biology Journal of the National Cancer Institute Molecular Vision Nature Publishing Group Nature Reviews Molecular Cell Biology Nature Genetics Nature Reviews Immunology Nature Reviews Cancer Nature Reviews Neuroscience Nature Biotechnology Nature Methods Neuro Oncology Placenta PLo. S ONE Society for Endocrinology journals Journal of Endocrinology Journal of Molecular Endocrinology Endocrine Related Cancer *Requirements vary with each journal.

Cell line authentication to improve reproducibility in cancer research NIH revised guidelines for funding – Enhancing Reproducibility through Rigor and Transparence (effective Jan 25, 2016) Authentication of key biological and/or chemical resources “NIH plans to enhance reproducibility in multiple ways, by promoting greater scientific rigor and transparency in funding applications and publications, encouraging robust peer review, providing adequate training, removing perverse incentives, and emphasizing overlooked areas such as the consideration of both sexes in research and the use of authenticated cell lines. ” Proprietary Information. Not for further distribution.

PURPOSE The contamination or misidentification of cell cultures can seriously compromise research performed with such cell lines. The purpose of this policy is to ensure that information generated using cultured cells at The University of MD Anderson Cancer Center (MD Anderson) is obtained from samples that have been authenticated by DNA validation techniques, to rule out misidentification and inter‐ or intra‐species contamination. This will prevent the necessity for retraction of papers in which cell lines are found not to be of the reported lineage. It has been indicated by NIH that grant applications that fail to use acceptable experimental practices “would not fare well in the review process. ” POLICY STATEMENT Validation by DNA analysis will be performed on cell lines used to produce scientific information at MD Anderson. This validation process will enhance the quality of research at MD Anderson and maintain the scientific integrity and reputation of the institution. SCOPE This policy applies to all workforce members utilizing cell lines for research. Proprietary Information. Not for further distribution.
New ANSI/ATCC Standards Development Organization Guidelines ASN 0002 consensus standard describing the use of short tandem repeat (STR) analysis for cell line authentication With STR profiling authentication may include: (i) verifying the cells are of human origin (ii) evaluating the consistency of profiles between isolates/passages (iii) comparing STR profile to a database (iv) detecting contaminating human DNA Proprietary Information. Not for further distribution.
Strengths & Limitations • STR systems can determine… Relatedness of a cell line to a reference standard Cell line cross contamination Gender (sex) • STR systems cannot distinguish… Different cell lines created from the same individual (donor) Cell lines created from identical twins • Promega STR systems are designed to be human specific Cannot be used to genotype non‐human species Proprietary Information. Not for further distribution.

Proprietary Information. Not for further distribution.
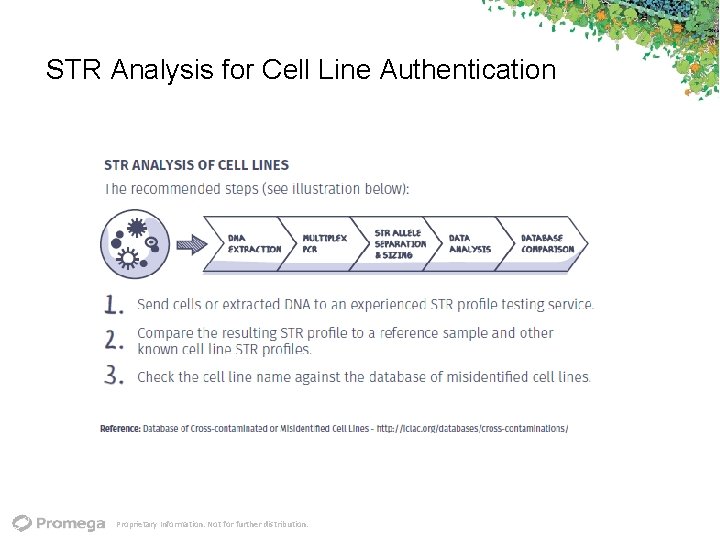
STR Analysis for Cell Line Authentication Proprietary Information. Not for further distribution.
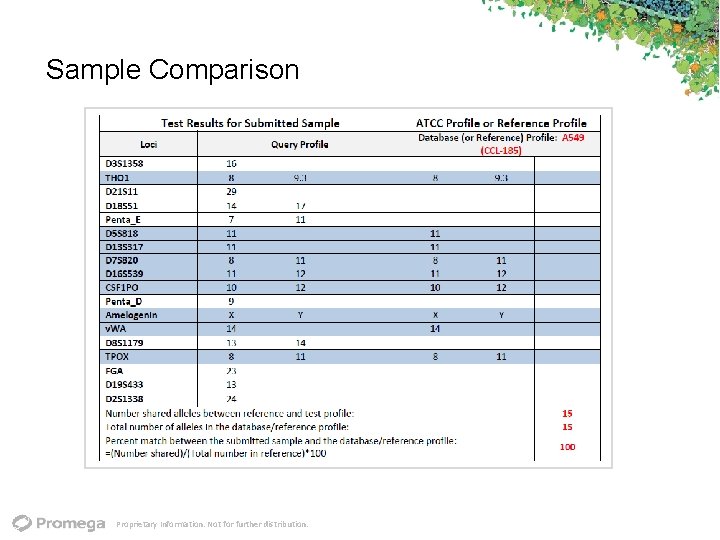
Sample Comparison Proprietary Information. Not for further distribution.
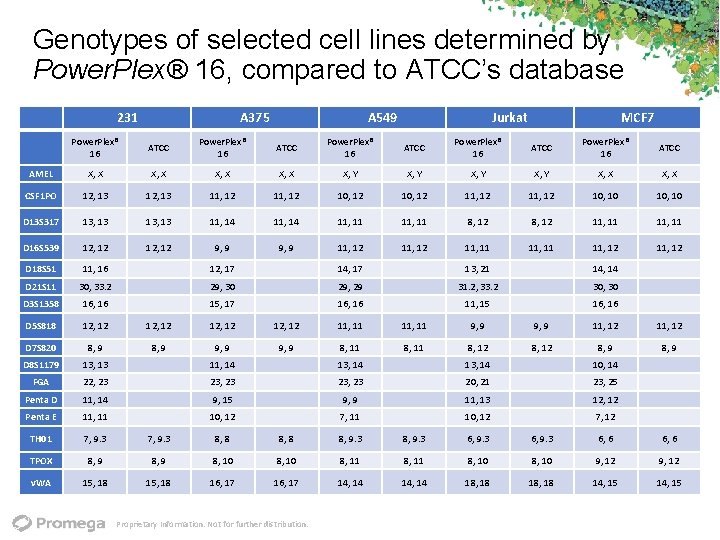
Genotypes of selected cell lines determined by Power. Plex® 16, compared to ATCC’s database 231 A 375 A 549 Jurkat MCF 7 Power. Plex® 16 ATCC AMEL X, X X, Y X, X CSF 1 PO 12, 13 11, 12 10, 12 11, 12 10, 10 D 13 S 317 13, 13 11, 14 11, 11 8, 12 11, 11 D 16 S 539 12, 12 9, 9 11, 12 11, 11 11, 12 D 18 S 51 11, 16 12, 17 14, 17 13, 21 14, 14 D 21 S 11 30, 33. 2 29, 30 29, 29 31. 2, 33. 2 30, 30 D 3 S 1358 16, 16 15, 17 16, 16 11, 15 16, 16 D 5 S 818 12, 12 11, 11 9, 9 11, 12 D 7 S 820 8, 9 9, 9 8, 11 8, 12 8, 9 D 8 S 1179 13, 13 11, 14 13, 14 10, 14 FGA 22, 23 23, 23 20, 21 23, 25 Penta D 11, 14 9, 15 9, 9 11, 13 12, 12 Penta E 11, 11 10, 12 7, 12 TH 01 7, 9. 3 8, 8 8, 9. 3 6, 6 TPOX 8, 9 8, 10 8, 11 8, 10 9, 12 v. WA 15, 18 16, 17 14, 14 18, 18 14, 15 Proprietary Information. Not for further distribution.
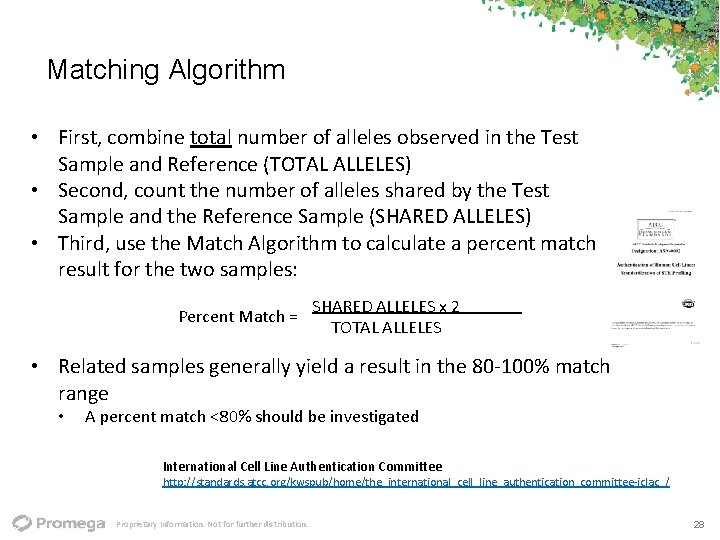
Matching Algorithm • First, combine total number of alleles observed in the Test Sample and Reference (TOTAL ALLELES) • Second, count the number of alleles shared by the Test Sample and the Reference Sample (SHARED ALLELES) • Third, use the Match Algorithm to calculate a percent match result for the two samples: Percent Match = SHARED ALLELES x 2 TOTAL ALLELES • Related samples generally yield a result in the 80‐ 100% match range • A percent match <80% should be investigated International Cell Line Authentication Committee http: //standards. atcc. org/kwspub/home/the_international_cell_line_authentication_committee‐iclac_/ Proprietary Information. Not for further distribution. 28
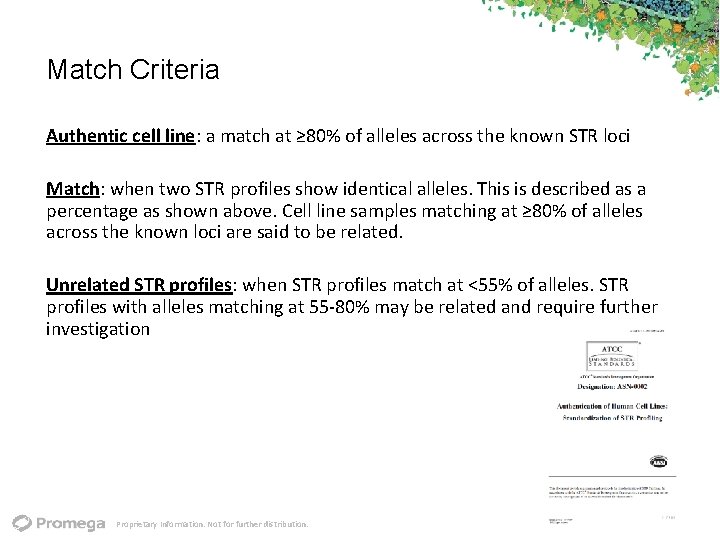
Match Criteria Authentic cell line: a match at ≥ 80% of alleles across the known STR loci Match: when two STR profiles show identical alleles. This is described as a percentage as shown above. Cell line samples matching at ≥ 80% of alleles across the known loci are said to be related. Unrelated STR profiles: when STR profiles match at <55% of alleles. STR profiles with alleles matching at 55‐ 80% may be related and require further investigation Proprietary Information. Not for further distribution.
Proprietary Information. Not for further distribution. https: //research. pathology. wisc. edu/cell‐line‐ authentication/

Sample Drop Off and Pricing Proprietary Information. Not for further distribution.
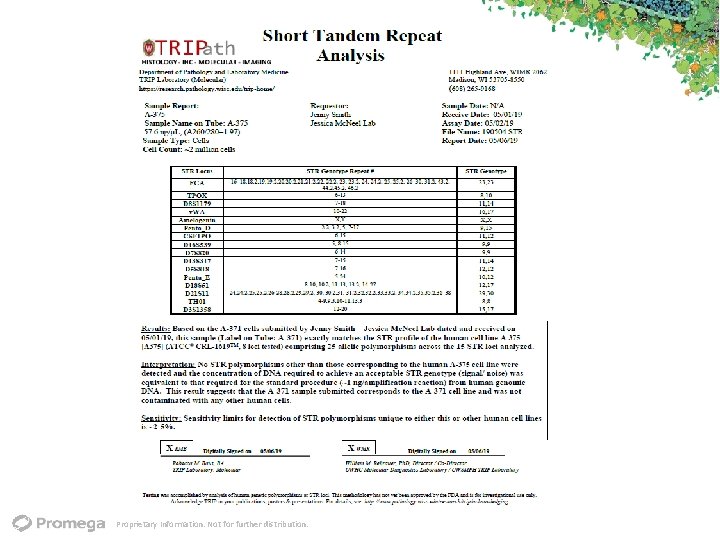
Proprietary Information. Not for further distribution.
Website: http: //www. promega. com/cla Proprietary Information. Not for further distribution.

Thank You! Nicole Sanderlin Field Support Scientist Nicole. Sanderlin@promega. com Technical Services Genetic@promega. com
- Slides: 34