ATOM ATOM ATOM ATOM ATOM ATOM ATOM ATOM










- Slides: 23





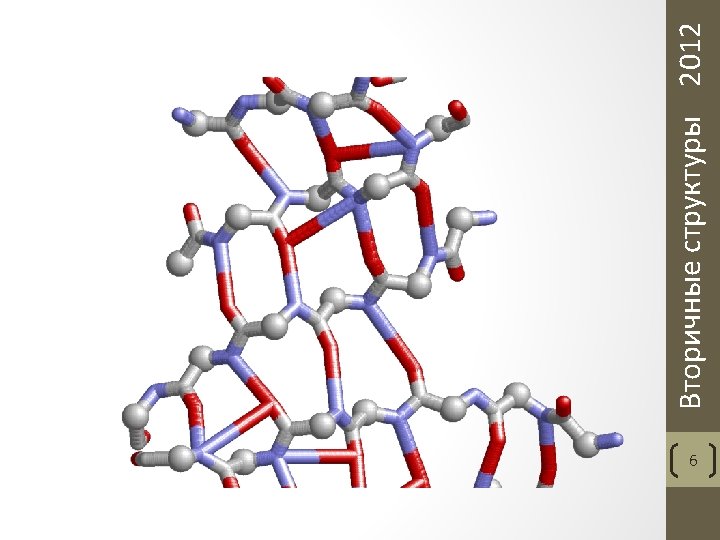
ATOM ATOM ATOM ATOM ATOM ATOM ATOM ATOM 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 N CA C O CB CG SD CE N CA C O CB CG CD CE NZ N CA C O CB CG CD 1 CD 2 CE 1 CE 2 CZ N CA MET MET LYS LYS LYS PHE PHE PHE GLY A A A A A A A A 1 1 1 1 2 2 2 2 2 3 3 3 4 4 65. 266 65. 196 63. 737 63. 070 66. 023 66. 327 67. 596 68. 730 63. 252 61. 878 61. 898 62. 848 61. 062 60. 901 60. 856 62. 203 62. 059 60. 814 60. 705 59. 358 58. 336 60. 960 62. 384 63. 412 62. 659 64. 734 63. 980 64. 988 59. 421 58. 224 8. 056 8. 000 8. 186 8. 989 9. 076 8. 769 7. 490 8. 059 7. 430 7. 545 8. 008 7. 691 6. 252 5. 819 4. 293 3. 593 2. 135 8. 741 9. 250 8. 945 9. 160 10. 785 11. 080 10. 949 11. 435 11. 202 11. 712 11. 569 8. 456 8. 129 4. 367 5. 806 6. 154 5. 509 6. 485 7. 909 8. 110 6. 902 7. 142 7. 562 8. 995 9. 728 7. 425 5. 961 5. 771 5. 508 5. 327 9. 341 10. 658 11. 298 10. 669 10. 678 10. 274 11. 211 8. 954 10. 844 8. 566 9. 519 12. 532 13. 322 Вторичные структуры 2012 Постановка задачи Задание: нарисовать вторичные структуры на доске 5




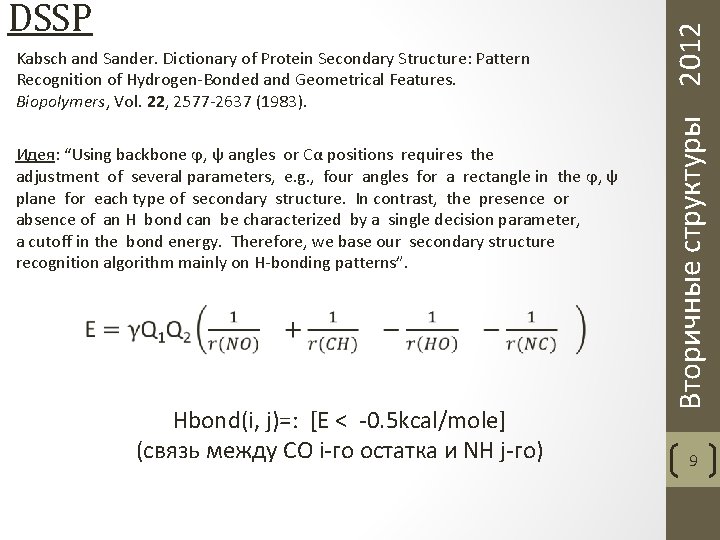
Kabsch and Sander. Dictionary of Protein Secondary Structure: Pattern Recognition of Hydrogen-Bonded and Geometrical Features. Biopolymers, Vol. 22, 2577 -2637 (1983). Идея: “Using backbone ϕ, ψ angles or Cα positions requires the adjustment of several parameters, e. g. , four angles for a rectangle in the ϕ, ψ plane for each type of secondary structure. In contrast, the presence or absence of an H bond can be characterized by a single decision parameter, a cutoff in the bond energy. Therefore, we base our secondary structure recognition algorithm mainly on H-bonding patterns”. Hbond(i, j)=: [E < -0. 5 kcal/mole] (связь между CO i-го остатка и NH j-го) Вторичные структуры 2012 DSSP 9

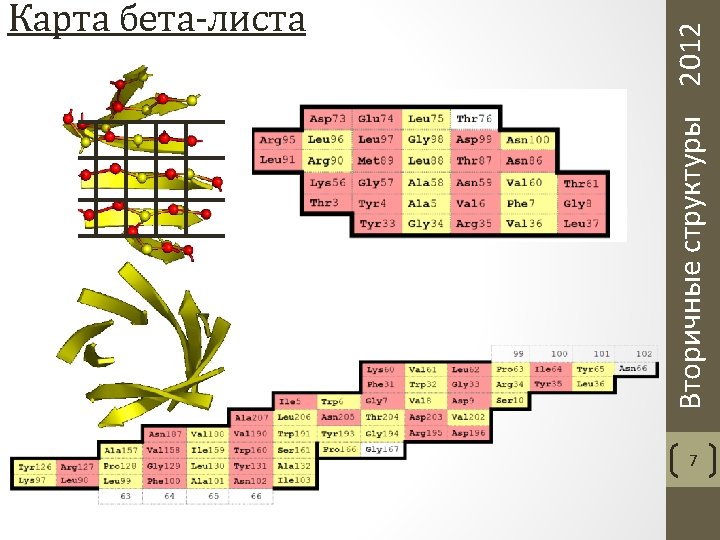
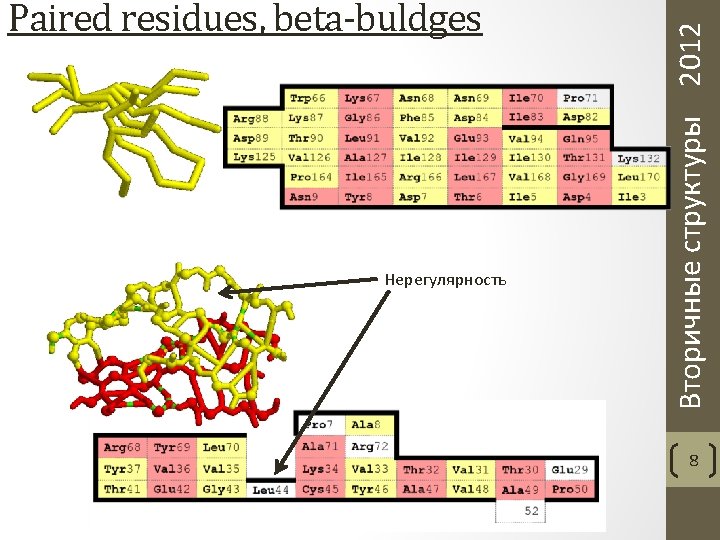
T: n-turn(i)=: Hbond(i, i+n), n = 3, 4, 5 H: 4 -helix(i, i+3) =: [4 -turn(i-1) and 4 -turn(i)] G: 3 -helix(i, i+2) =: [3 -turn(i-1) and 3 -turn(i)] I: 5 -helix(i, i+4) =: [5 -turn(i-1) and 5 -turn(i)] B: Parallel Bridge(i, j) =: [Hbond(i-1, j) and Hbond(j, i+1)] Antiparallel Bridge(i, j) =: [Hbond(i, j) and Hbond(j, i)] or [Hbond(i-1, j+1) and Hbond(j-1, i+1)] E: ladder =: set of one or more consecutive bridges of identical type sheet =: set of one or more ladders connected by shared residues Приоритет: H, B, E, G, I, T Вторичные структуры 2012 Паттерны DSSP 10

1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 A A A A A A A M D Q F E C I N V A D A H Q K L Q E K E A V L V D I S E E E H H H H H T T E E + S-a +a -a > > S+ X S+ <>S+ ><5 S+ 3<5 S< 5 S+ < + -b -b -bc + 0 0 87 88 89 0 0 0 0 61 62 63 64 0 0 0 A 0 A 0 A 0 0 0 0 A 0 A 41 A 42 A 0 102 135 128 46 94 61 9 81 2 48 54 0 42 108 32 13 136 88 191 100 8 45 3 0 0 41 0, 0. 0 83, -0. 0 81, -0. 2 -2, -0. 3 81, -1. 8 -2, -0. 4 81, -2. 4 -2, -0. 3 81, -0. 3 2, -0. 2 -4, -2. 6 -4, -1. 9 -4, -2. 4 -4, -2. 6 -4, -2. 2 -3, -1. 6 -5, -2. 6 -6, -0. 8 38, -2. 7 16, -0. 5 38, -2. 9 16, -2. 8 38, -2. 3 85, -0. 1 2, -0. 3 83, -0. 3 2, -0. 4 83, -2. 2 2, -0. 3 83, -2. 8 4, -2. 6 4, -1. 9 4, -2. 4 4, -2. 6 4, -2. 2 5, -2. 6 3, -1. 6 -1, -0. 2 -1, -0. 3 -3, -0. 2 2, -0. 3 40, -2. 7 18, -2. 5 40, -2. 7 18, -1. 9 39, -0. 1 Вторичные структуры 2012 DSSP 0, 0. 0 2, -0. 0 76, -0. 0 81, -0. 1 -2, -0. 3 81, -0. 2 -2, -0. 5 81, -0. 2 2, -0. 2 1, -0. 2 -5, -0. 2 1, -0. 3 -5, -0. 2 -4, -0. 3 -6, -0. 2 -9, -0. 1 -2, -0. 3 11 -2, -0. 3 -2, -0. 5 -2, -0. 4 -2, -0. 5



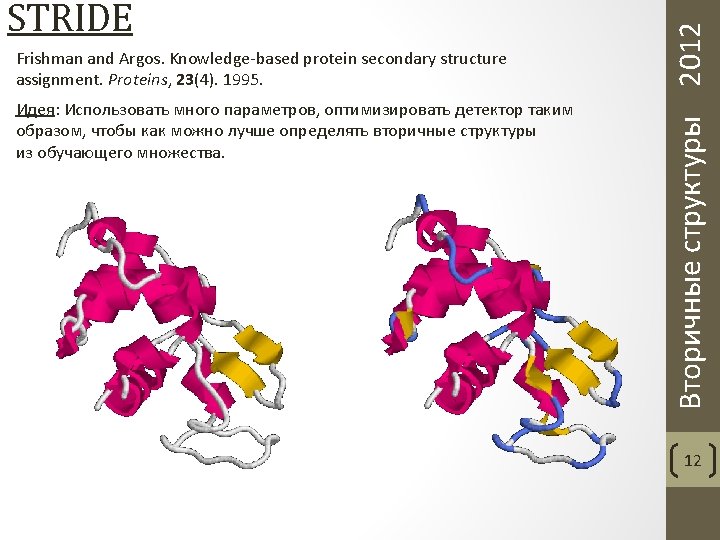
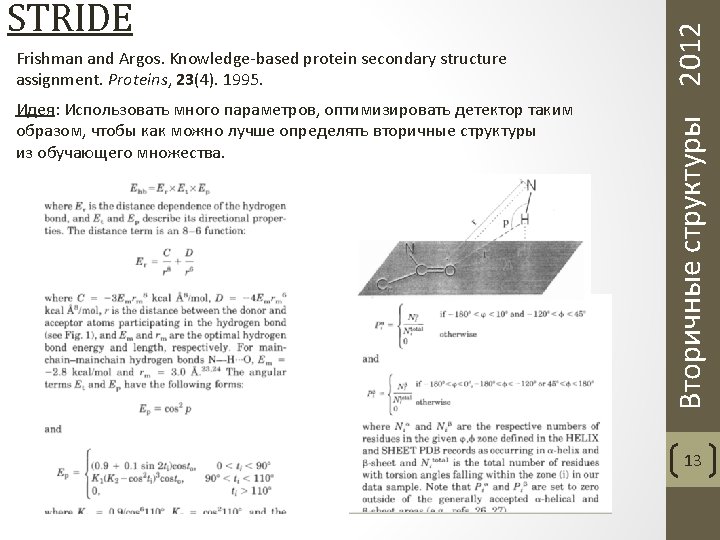
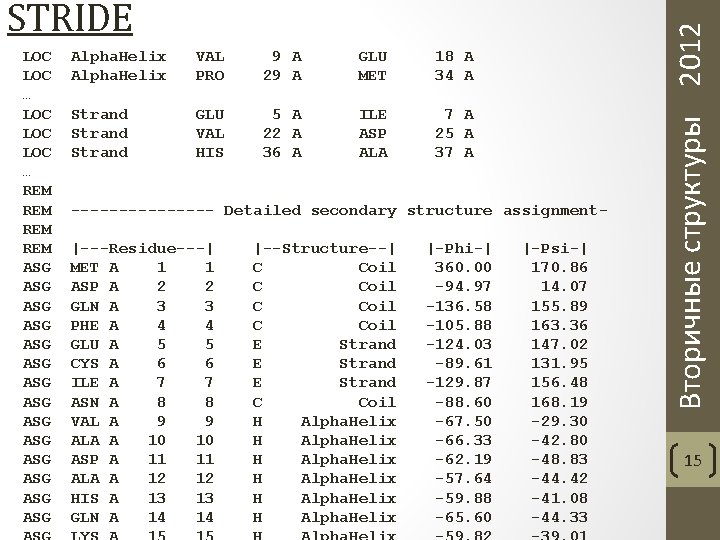
Frishman and Argos. Knowledge-based protein secondary structure assignment. Proteins, 23(4). 1995. Вторичные структуры 2012 STRIDE 14

LOC … LOC LOC … REM REM ASG ASG ASG ASG Alpha. Helix VAL PRO 9 A 29 A GLU MET 18 A 34 A Strand GLU VAL HIS 5 A 22 A 36 A ILE ASP ALA 7 A 25 A 37 A -------- Detailed secondary structure assignment|---Residue---| MET A 1 1 ASP A 2 2 GLN A 3 3 PHE A 4 4 GLU A 5 5 CYS A 6 6 ILE A 7 7 ASN A 8 8 VAL A 9 9 ALA A 10 10 ASP A 11 11 ALA A 12 12 HIS A 13 13 GLN A 14 14 |--Structure--| C Coil E Strand C Coil H Alpha. Helix |-Phi-| 360. 00 -94. 97 -136. 58 -105. 88 -124. 03 -89. 61 -129. 87 -88. 60 -67. 50 -66. 33 -62. 19 -57. 64 -59. 88 -65. 60 |-Psi-| 170. 86 14. 07 155. 89 163. 36 147. 02 131. 95 156. 48 168. 19 -29. 30 -42. 80 -48. 83 -44. 42 -41. 08 -44. 33 Вторичные структуры 2012 STRIDE 15

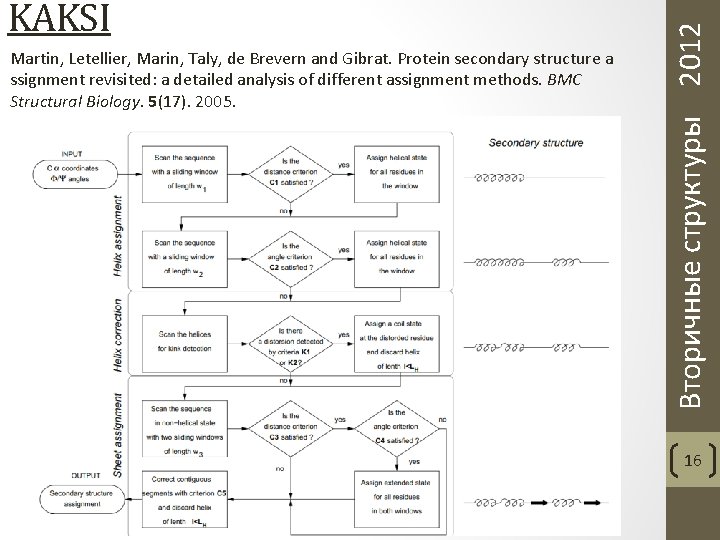
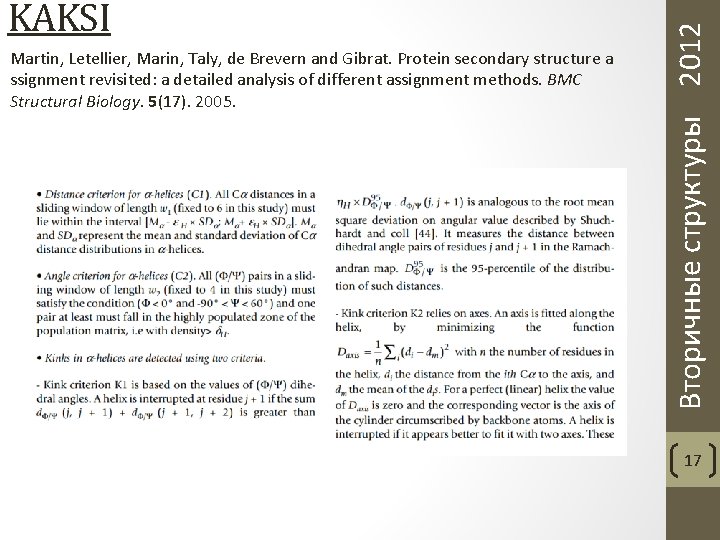
Martin, Letellier, Marin, Taly, de Brevern and Gibrat. Protein secondary structure a ssignment revisited: a detailed analysis of different assignment methods. BMC Structural Biology. 5(17). 2005. Вторичные структуры 2012 KAKSI 16

Martin, Letellier, Marin, Taly, de Brevern and Gibrat. Protein secondary structure a ssignment revisited: a detailed analysis of different assignment methods. BMC Structural Biology. 5(17). 2005. Вторичные структуры 2012 KAKSI 17

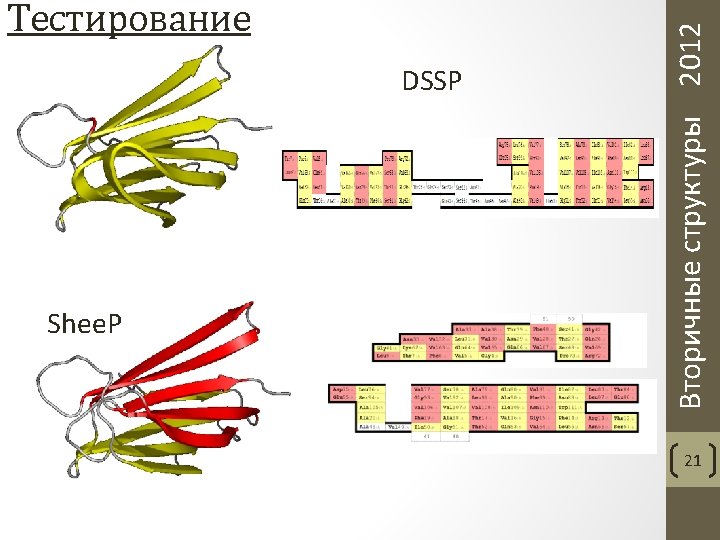
Вторичные структуры 2012 Sheep: Sheet Puzzle 18


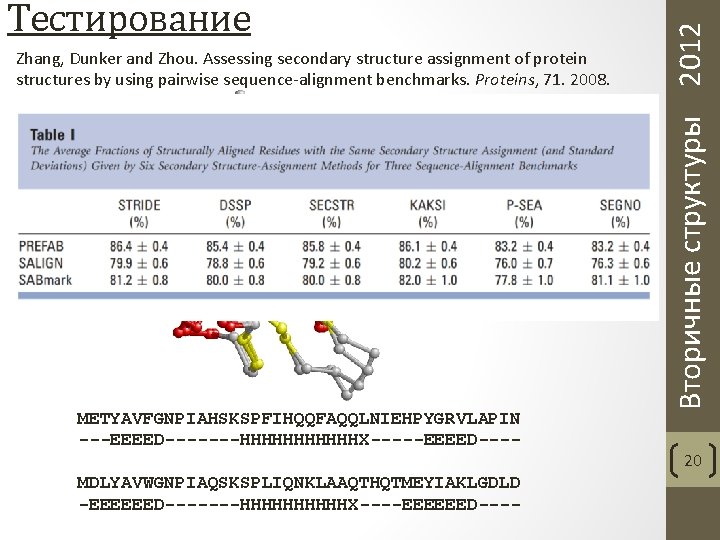
Zhang, Dunker and Zhou. Assessing secondary structure assignment of protein structures by using pairwise sequence-alignment benchmarks. Proteins, 71. 2008. METYAVFGNPIAHSKSPFIHQQFAQQLNIEHPYGRVLAPIN ---EEEED-------HHHHHHX-----EEEED---MDLYAVWGNPIAQSKSPLIQNKLAAQTHQTMEYIAKLGDLD -EEEEEED-------HHHHHX----EEEEEED---- Вторичные структуры 2012 Тестирование 20


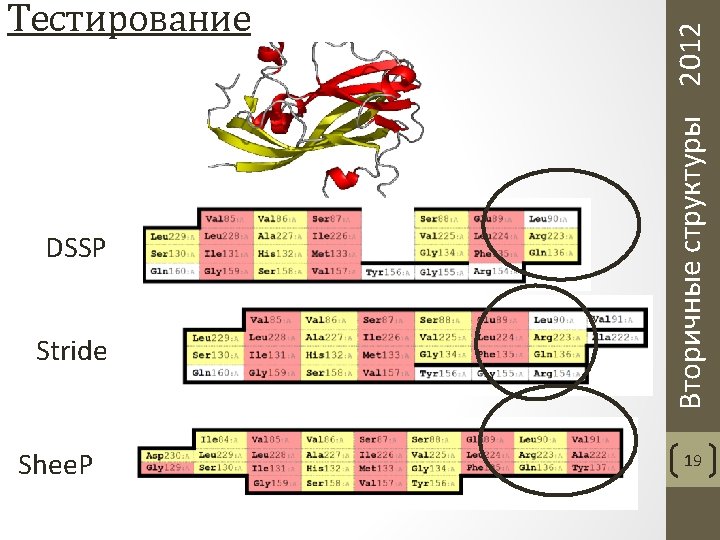
Частота ошибок, допускаемых программами DSSP, Stride и Shee. P Aksianov, Alexeevski. Shee. P: A Tool for Description of β-Sheets in Protein 3 D Structures. Journal of Bioinformatics and Computational Biology, 2012. 10(2) Вторичные структуры 2012 Тестирование 22
