At the end of the electron transport chain


![High [H+] + + + High [H+] Inner Membrane Matrix + + ++ + High [H+] + + + High [H+] Inner Membrane Matrix + + ++ +](https://slidetodoc.com/presentation_image/1995fae42244b8df4a8a5e9259217459/image-10.jpg)



- Slides: 47

At the end of the electron transport chain, oxygen receives the energy-spent electrons, resulting in the production of water. ½ O 2 + 2 e- + 2 H+ → H 2 O Oxygen is the final electron acceptor. Almost immediately oxidized into H 2 O

How is this coupling accomplished? was originally thought that ATP generation was somehow directly done at Complexes I, III and IV. We now know that the coupling is indirect in that a proton gradient is generated across the inner mitochondrial membrane which drives ATP synthesis.

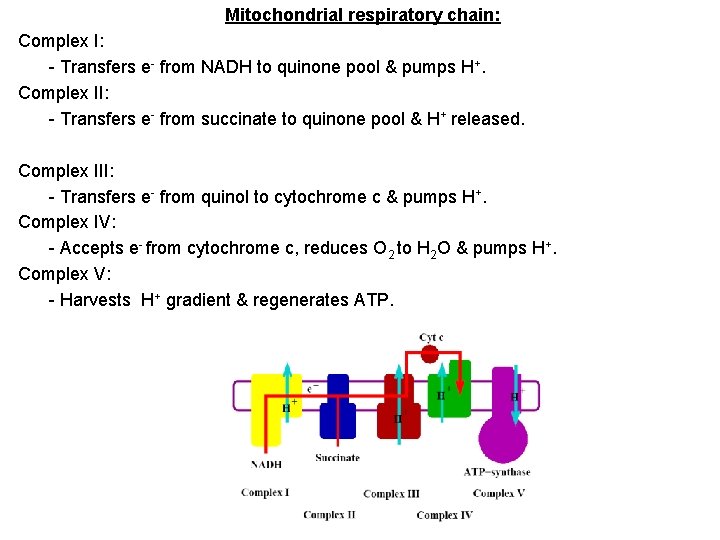
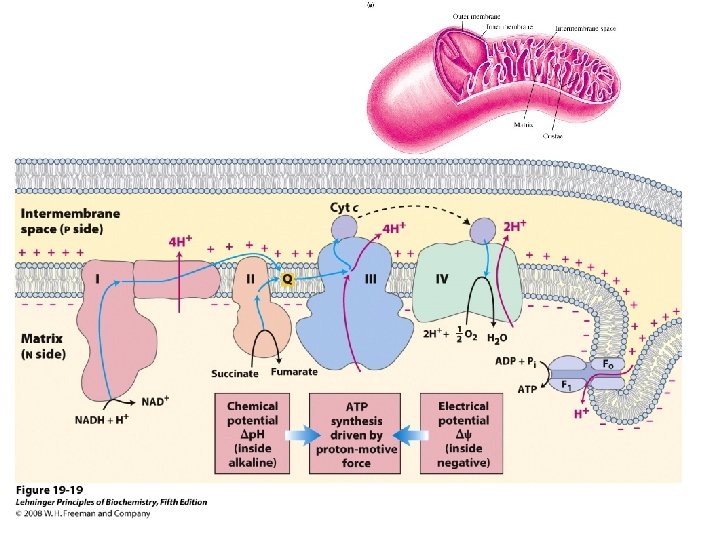
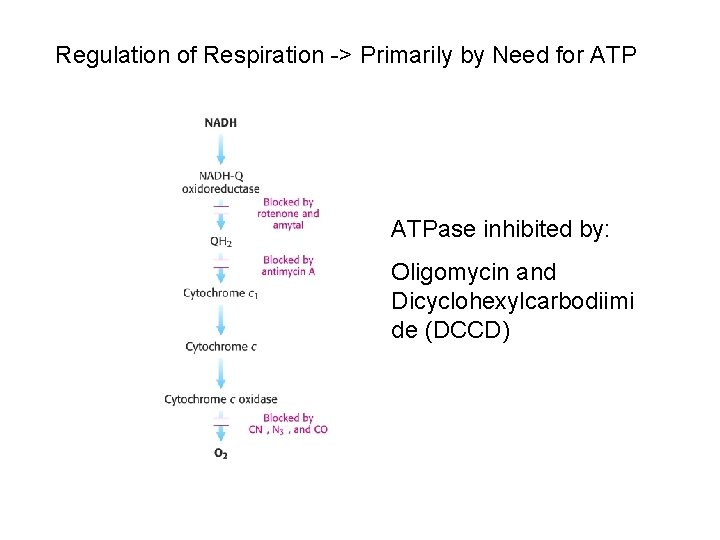
Mitochondrial respiratory chain: Complex I: - Transfers e- from NADH to quinone pool & pumps H+. Complex II: - Transfers e- from succinate to quinone pool & H+ released. Complex III: - Transfers e- from quinol to cytochrome c & pumps H+. Complex IV: - Accepts e- from cytochrome c, reduces O 2 to H 2 O & pumps H+. Complex V: - Harvests H+ gradient & regenerates ATP.

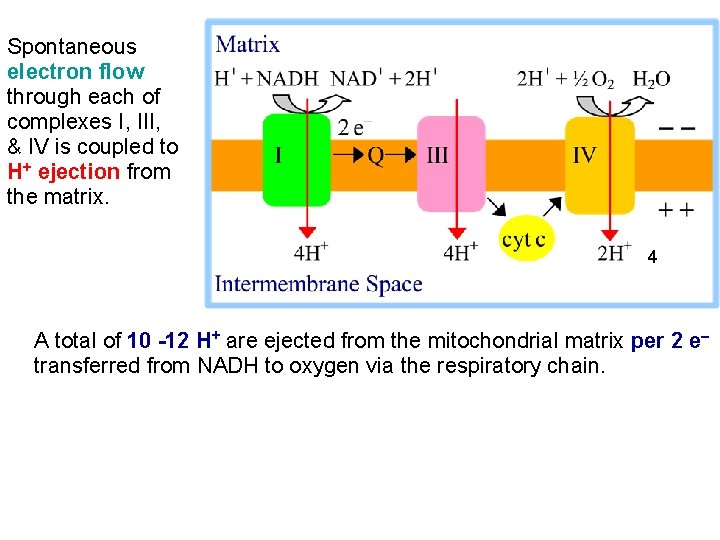
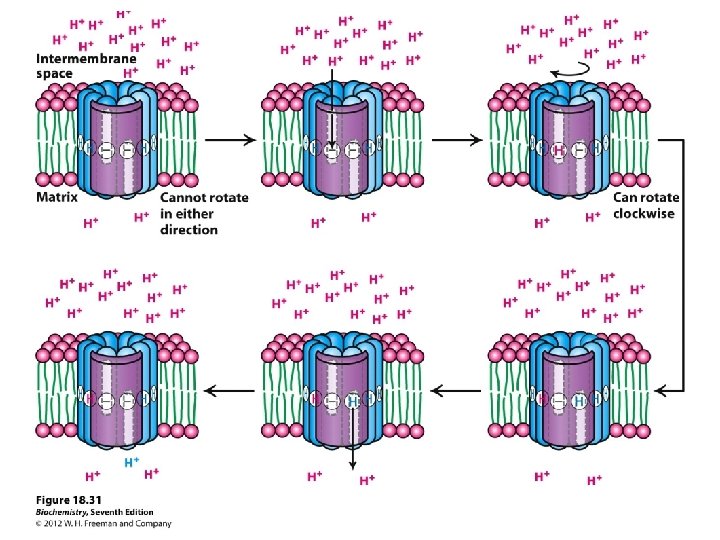
Spontaneous electron flow through each of complexes I, III, & IV is coupled to H+ ejection from the matrix. 4 A total of 10 -12 H+ are ejected from the mitochondrial matrix per 2 e- transferred from NADH to oxygen via the respiratory chain.

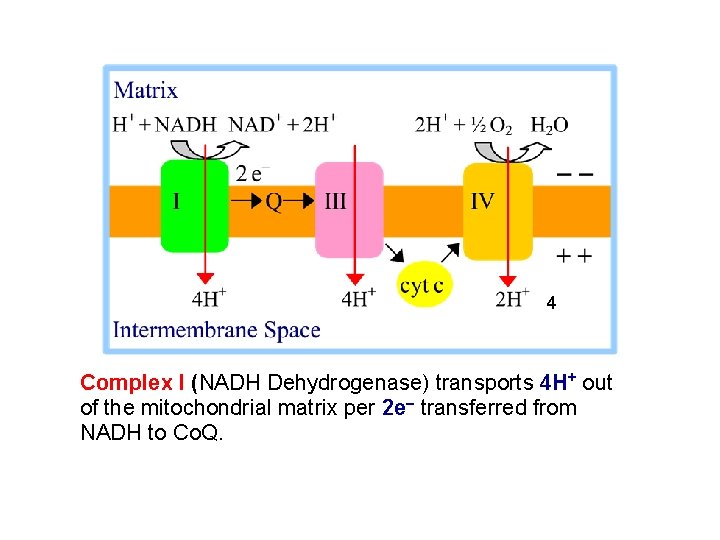
4 Complex I (NADH Dehydrogenase) transports 4 H+ out of the mitochondrial matrix per 2 e- transferred from NADH to Co. Q.

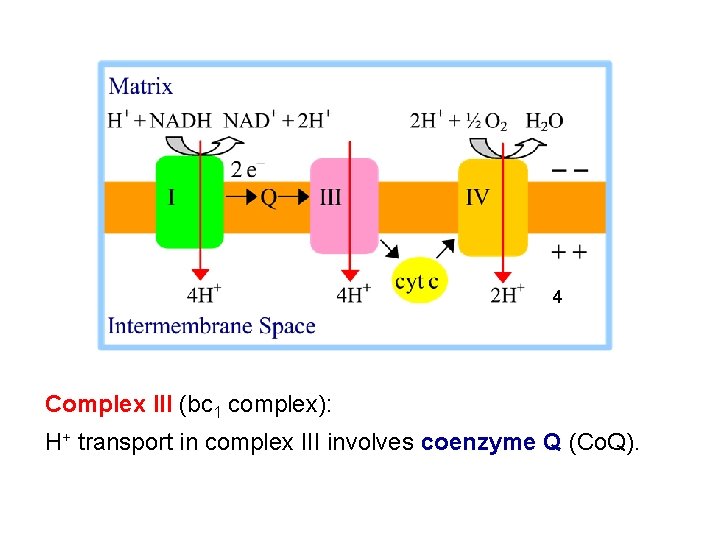
4 Complex III (bc 1 complex): H+ transport in complex III involves coenzyme Q (Co. Q).



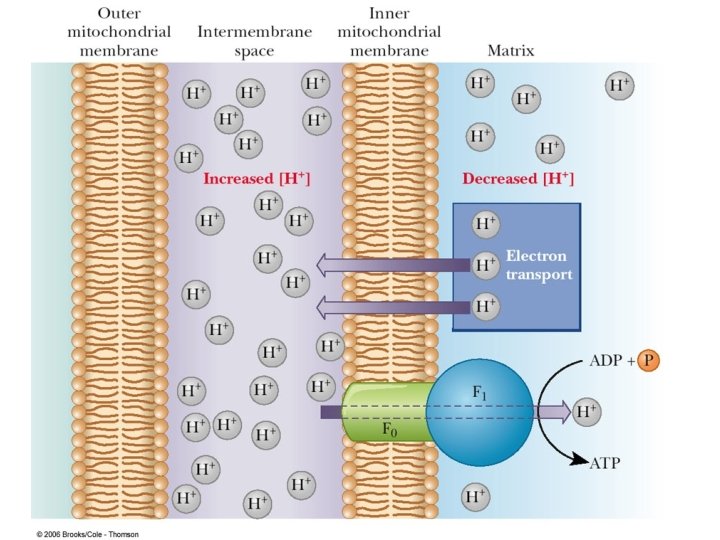
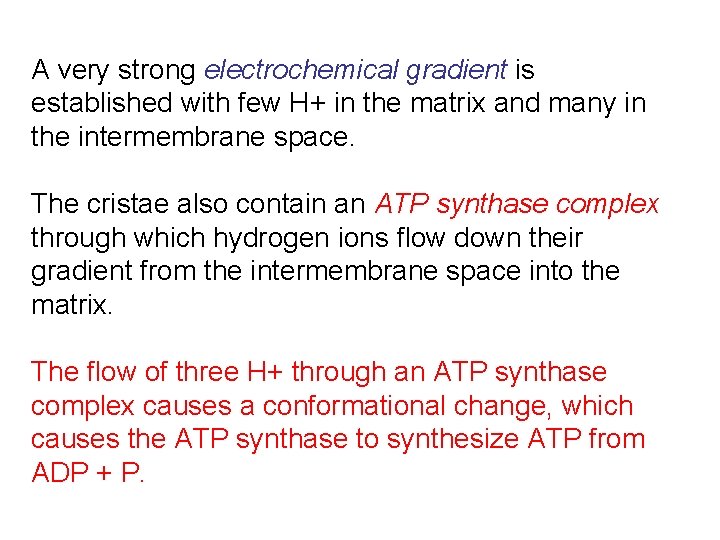
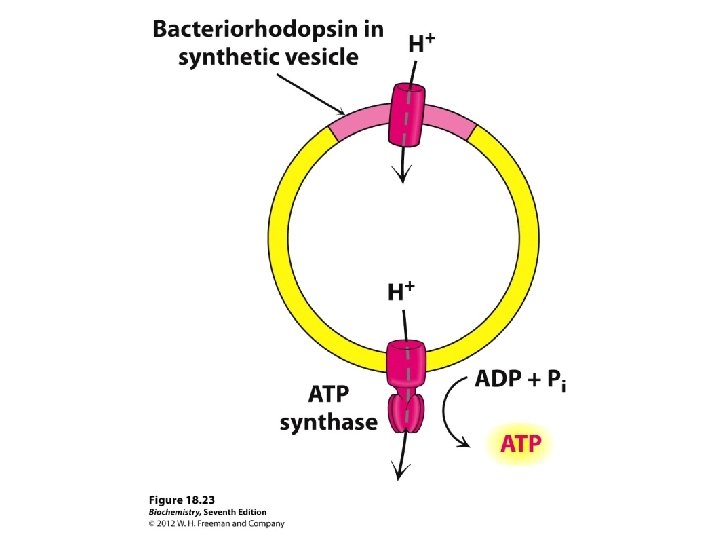
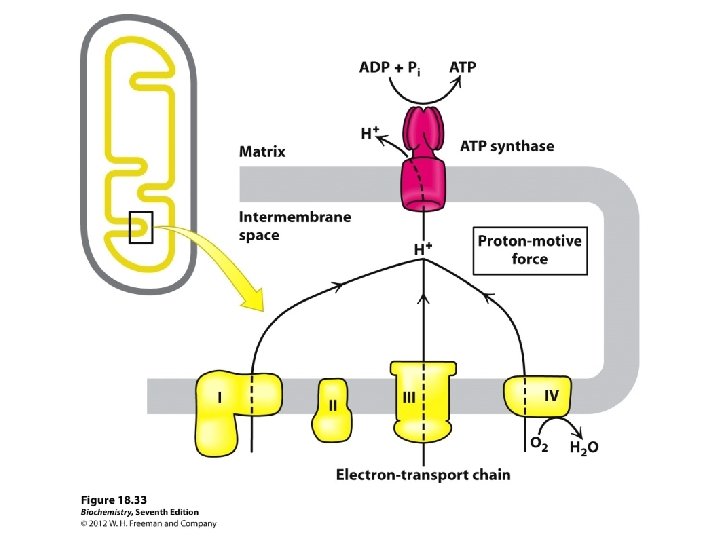
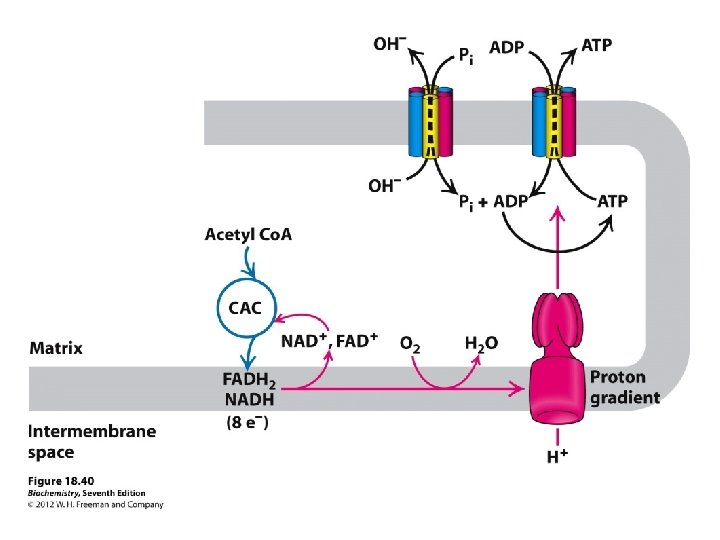
A very strong electrochemical gradient is established with few H+ in the matrix and many in the intermembrane space. The cristae also contain an ATP synthase complex through which hydrogen ions flow down their gradient from the intermembrane space into the matrix. The flow of three H+ through an ATP synthase complex causes a conformational change, which causes the ATP synthase to synthesize ATP from ADP + P.
![High H High H Inner Membrane Matrix High [H+] + + + High [H+] Inner Membrane Matrix + + ++ +](https://slidetodoc.com/presentation_image/1995fae42244b8df4a8a5e9259217459/image-10.jpg)
High [H+] + + + High [H+] Inner Membrane Matrix + + ++ + H+ + - H+ _ H+ + H _ H H+ Low [H+] H+ + H+ H Cytoplasm Outer Membrane Intermembrane Space + + + Cristae Generation of a p. H gradient ([H+]) and charge difference (negative in the matrix) across the inner membrane constitute the protonmotive force that can be used to drive ATP synthesis and transport processes.

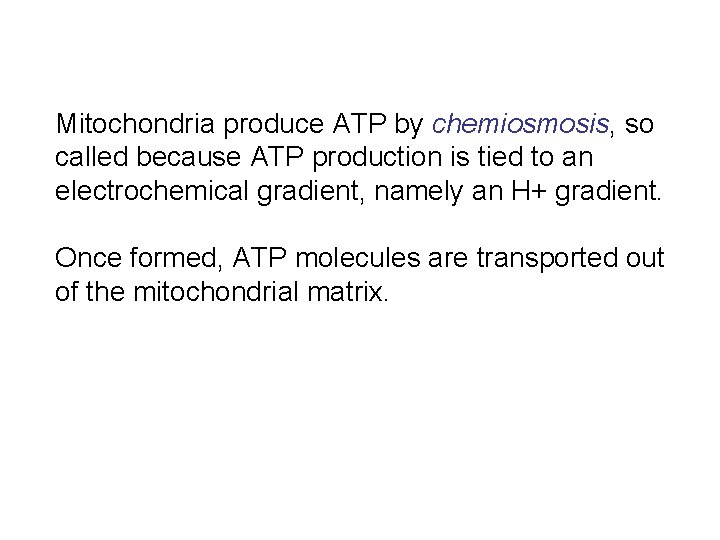
Mitochondria produce ATP by chemiosmosis, so called because ATP production is tied to an electrochemical gradient, namely an H+ gradient. Once formed, ATP molecules are transported out of the mitochondrial matrix.

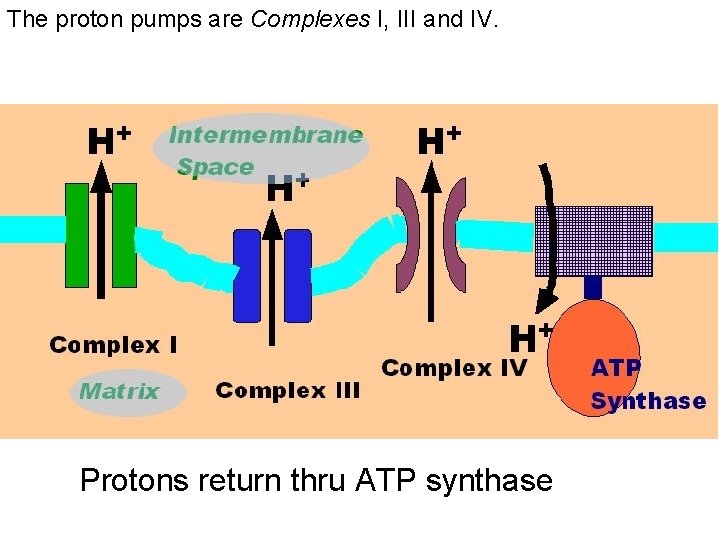
Chemiosmotic Theory --Peter Mitchell A proton gradient is generated with energy from electron transport by proton pumping by Complexes I, III, IV from the matrix to intermembrane space of the mitochondrion.

The protons have a thermodynamic tendency to return to the matrix = Proton-motive force The protons diffuse back into the matrix through the Fo. F 1 ATP synthase complex. The free energy release drives ATP synthesis.


The proton pumps are Complexes I, III and IV. Protons return thru ATP synthase


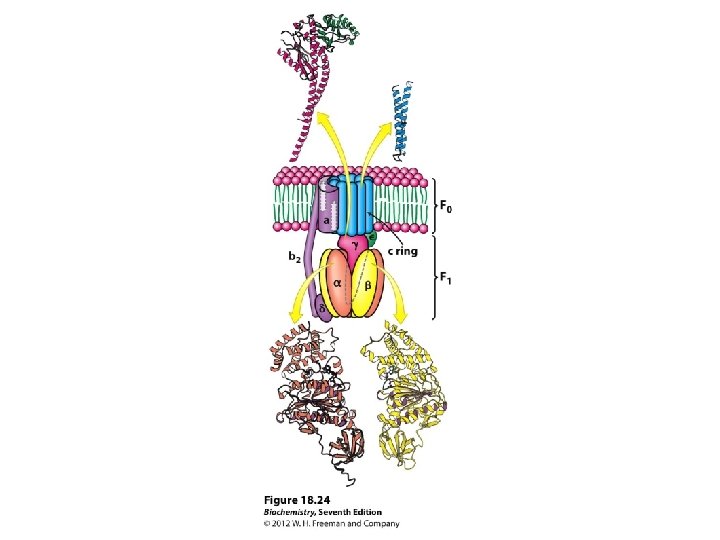
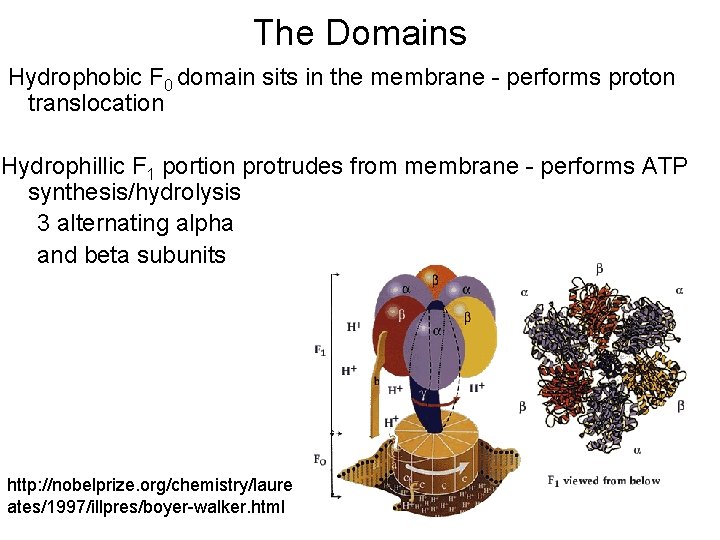
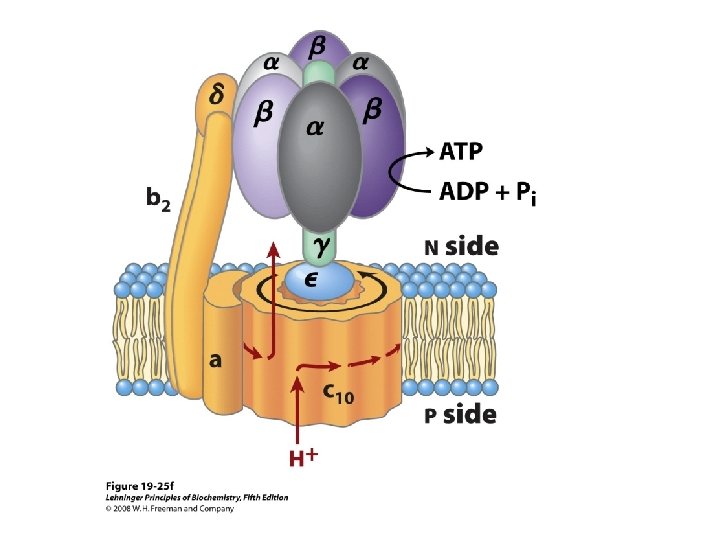
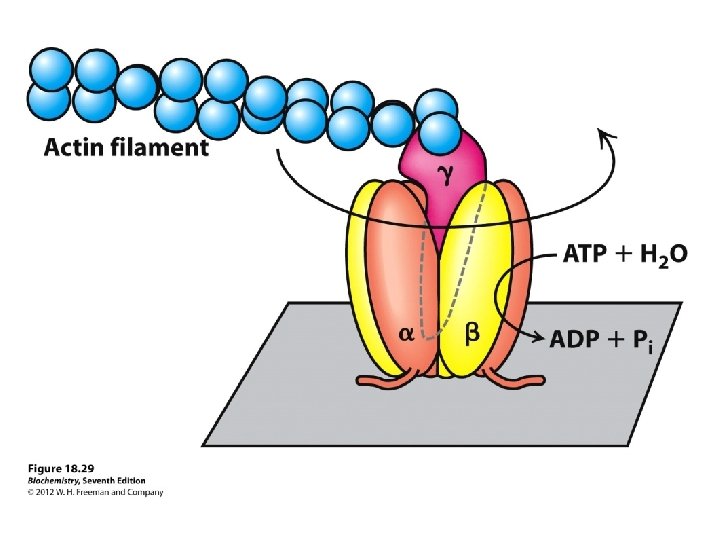
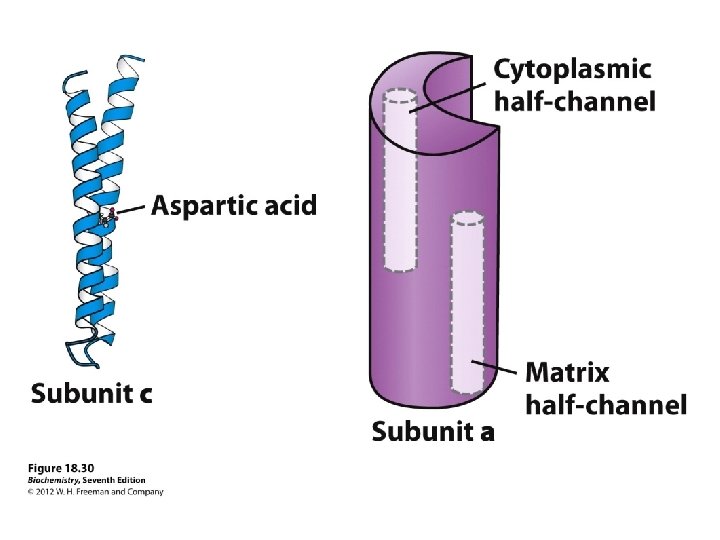
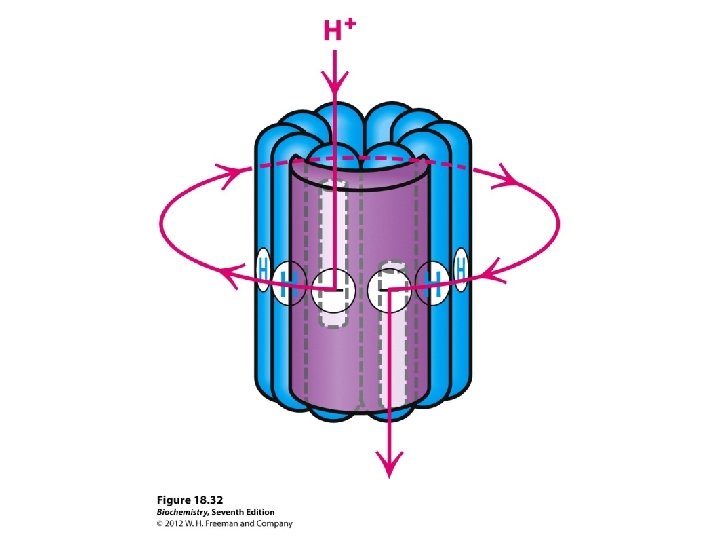
The Domains Hydrophobic F 0 domain sits in the membrane - performs proton translocation Hydrophillic F 1 portion protrudes from membrane - performs ATP synthesis/hydrolysis 3 alternating alpha and beta subunits http: //nobelprize. org/chemistry/laure ates/1997/illpres/boyer-walker. html


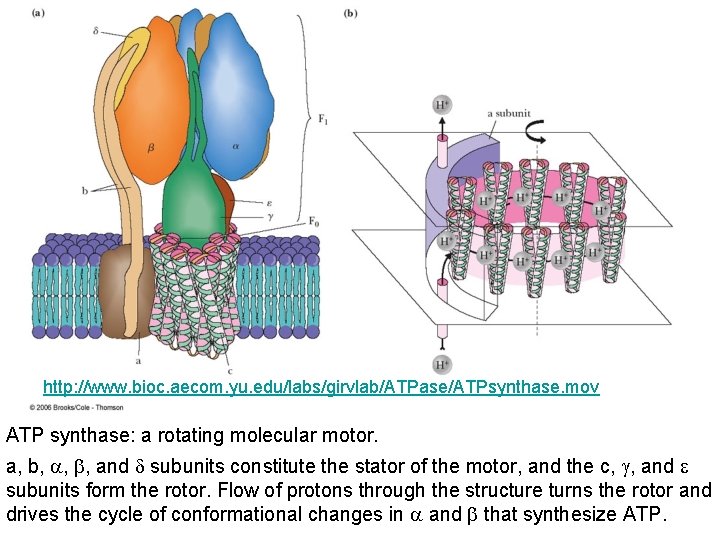
http: //www. bioc. aecom. yu. edu/labs/girvlab/ATPase/ATPsynthase. mov ATP synthase: a rotating molecular motor. a, b, , , and subunits constitute the stator of the motor, and the c, , and subunits form the rotor. Flow of protons through the structure turns the rotor and drives the cycle of conformational changes in and that synthesize ATP.

Animations http: //www. stolaf. edu/people/giannini/flashanimat/metabolism/atps yn 1. swf Protons cross membrane through the ATP synthase enzyme http: //www. stolaf. edu/people/giannini/flashanimat/metabolism/atps yn 2. swf Rotary motion of ATP synthase powers the synthesis of ATP

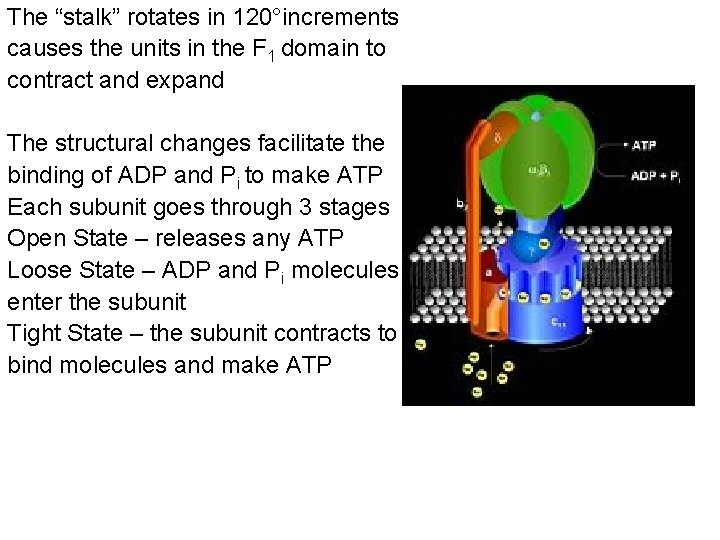
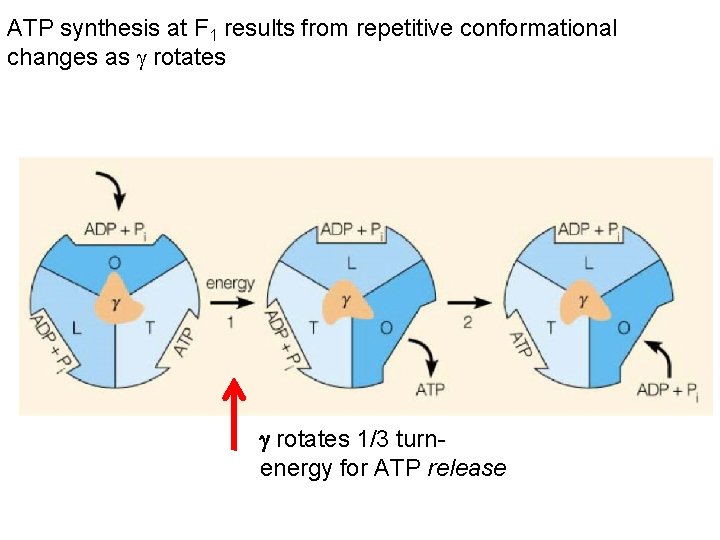
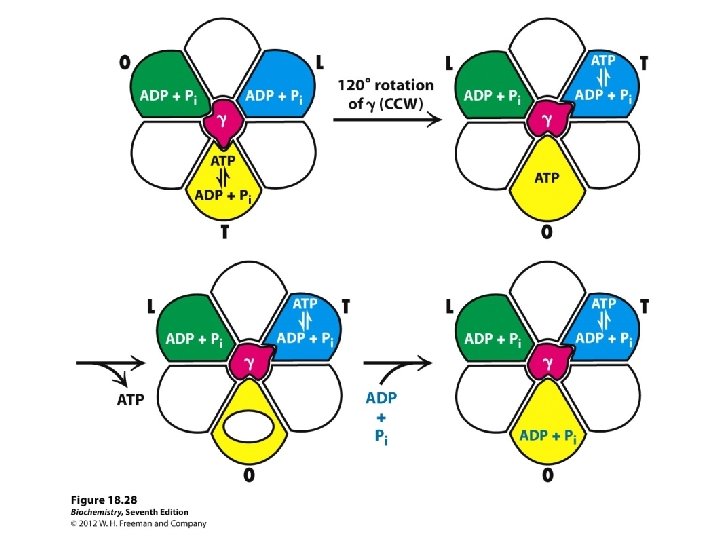
The “stalk” rotates in 120°increments causes the units in the F 1 domain to contract and expand The structural changes facilitate the binding of ADP and Pi to make ATP Each subunit goes through 3 stages Open State – releases any ATP Loose State – ADP and Pi molecules enter the subunit Tight State – the subunit contracts to bind molecules and make ATP

ATP synthesis at F 1 results from repetitive conformational changes as rotates 1/3 turn energy for ATP release








Interesting Facts Contains 22722 atoms 23211 bonds connected as 2987 amino acid groups 120 helix units and 94 sheet units Generates over 100 kg of ATP daily (in humans) One of the oldest enzymes-appeared earlier then photosynthetic or respiratory enzymes Smallest rotary machine known

Uncoupling. The compound 2, 4 dinitrophenol (DNP) allows H+ through the membrane and uncouples. Blocking. The antibiotic oligomycin blocks the flow of H+ through the Fo, directly inhibiting ox-phos.

Regulation of Respiration -> Primarily by Need for ATPase inhibited by: Oligomycin and Dicyclohexylcarbodiimi de (DCCD)

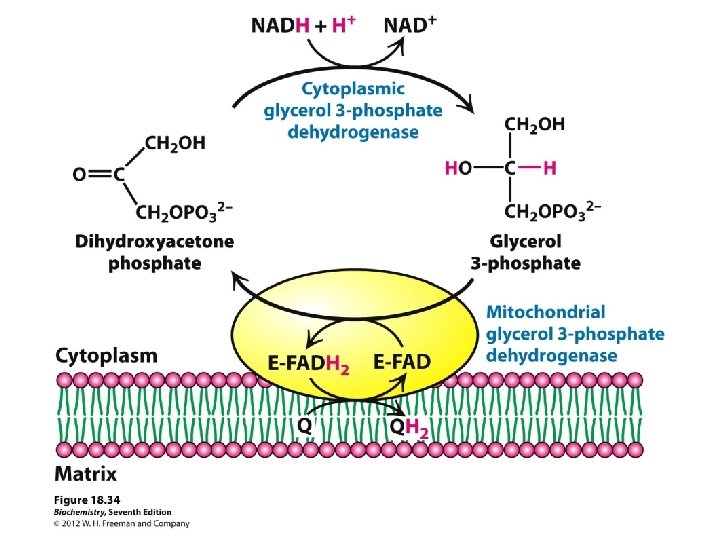
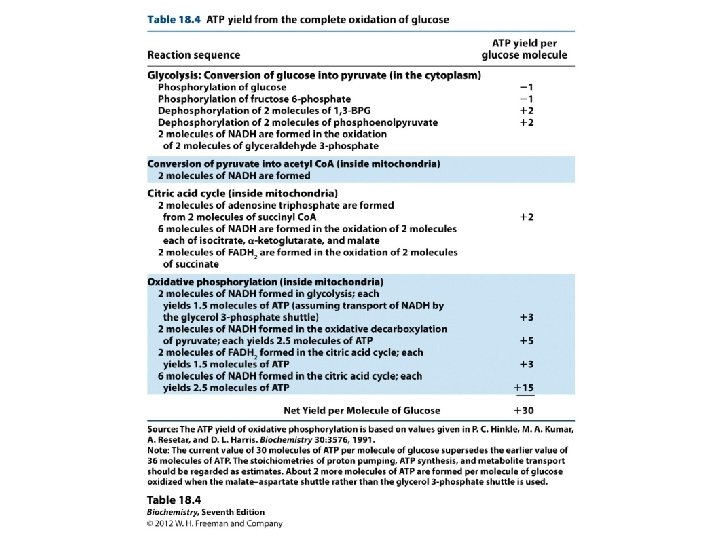
Chemiosmosis Production of ATP in Electron Transport H+ (Protons) generated from NADH Electrochemical Gradient Formed between membranes Electrical Force (+) & p. H Force (Acid) thus gradient formed ATPase enzyme that channels H+ from High to Low concentration 3 ATP/NADH 2 ATP/NADH




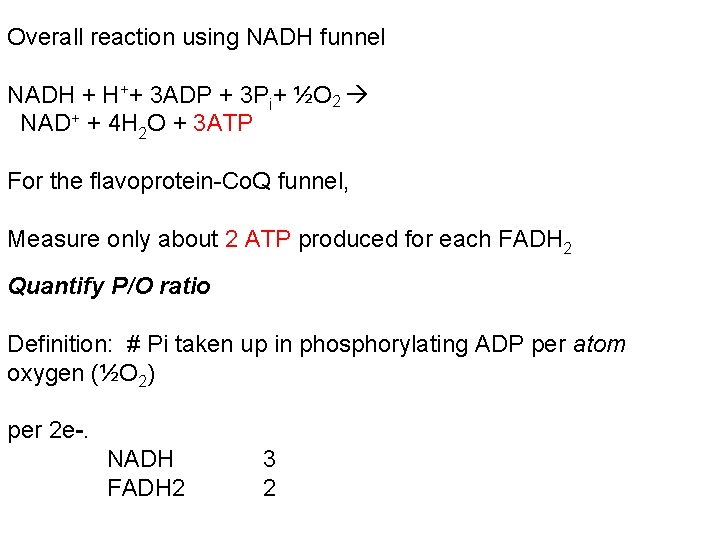
Overall reaction using NADH funnel NADH + H++ 3 ADP + 3 Pi+ ½O 2 NAD+ + 4 H 2 O + 3 ATP For the flavoprotein-Co. Q funnel, Measure only about 2 ATP produced for each FADH 2 Quantify P/O ratio Definition: # Pi taken up in phosphorylating ADP per atom oxygen (½O 2) per 2 e-. NADH FADH 2 3 2

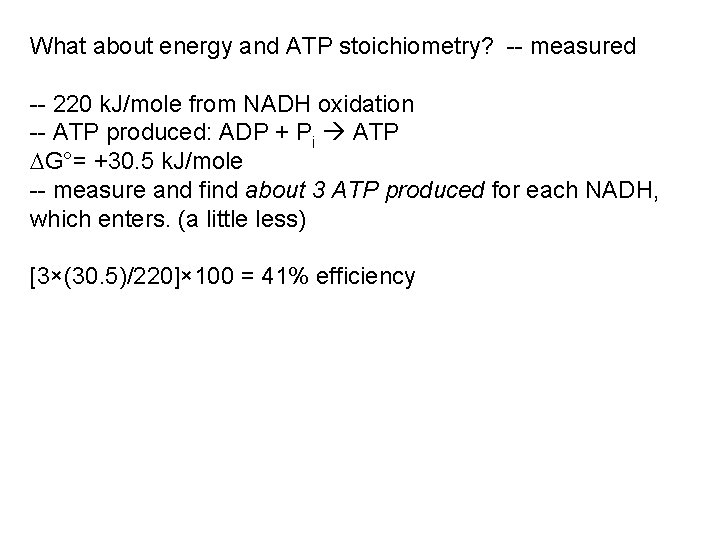
What about energy and ATP stoichiometry? -- measured -- 220 k. J/mole from NADH oxidation -- ATP produced: ADP + Pi ATP G°= +30. 5 k. J/mole -- measure and find about 3 ATP produced for each NADH, which enters. (a little less) [3×(30. 5)/220]× 100 = 41% efficiency

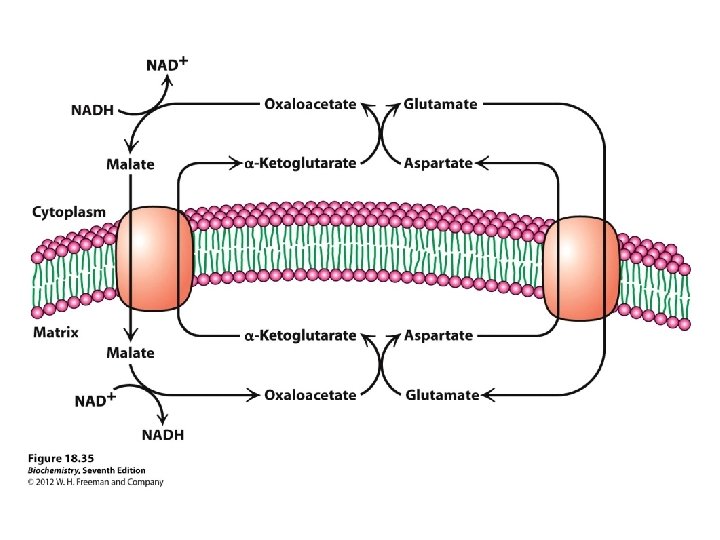
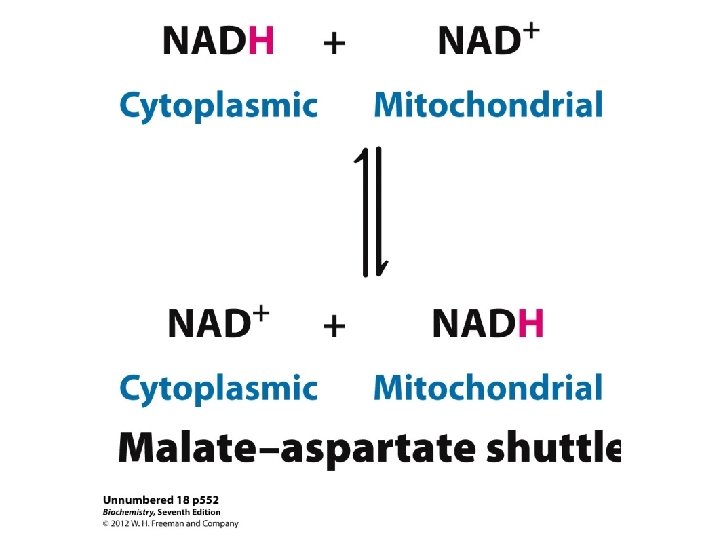
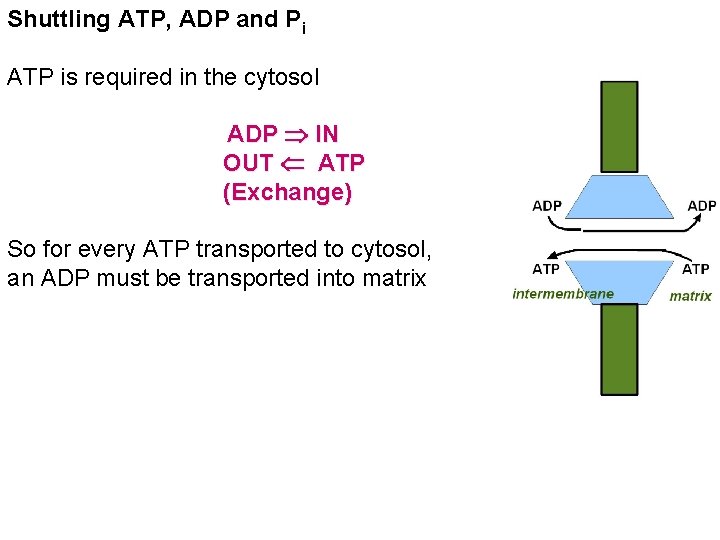
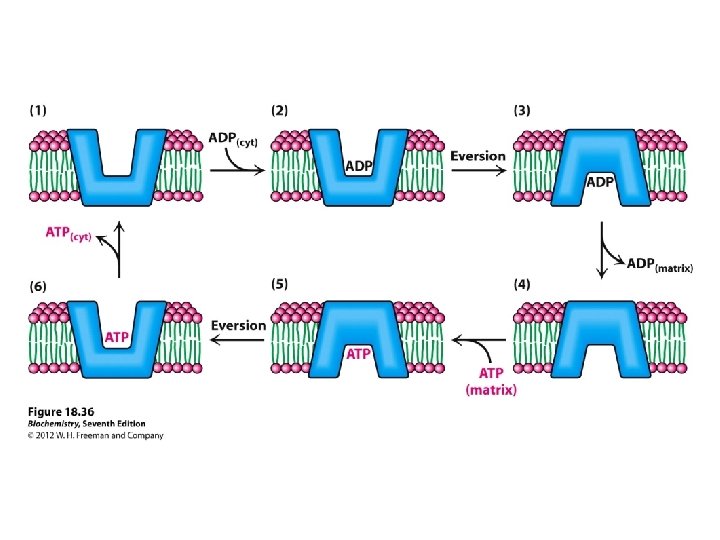
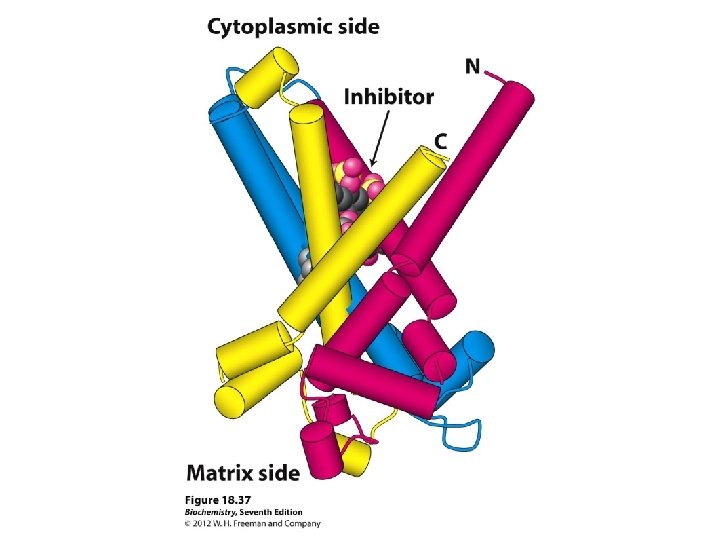
Shuttling ATP, ADP and Pi ATP is required in the cytosol ADP IN OUT ATP (Exchange) So for every ATP transported to cytosol, an ADP must be transported into matrix


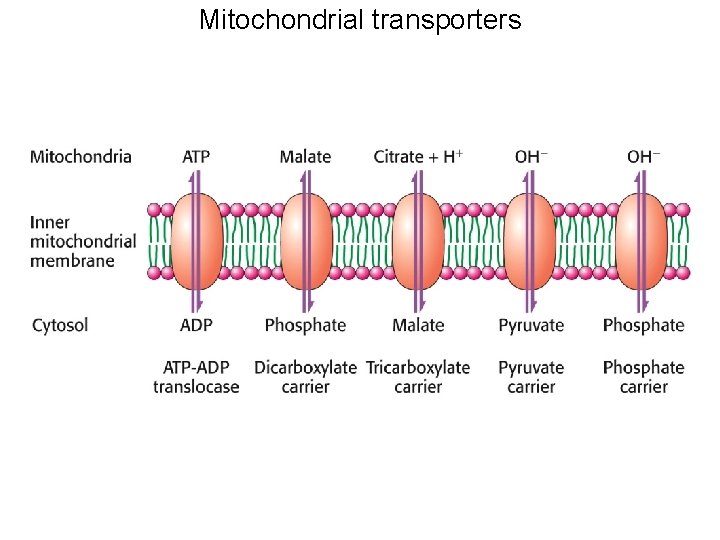
Mitochondrial transporters

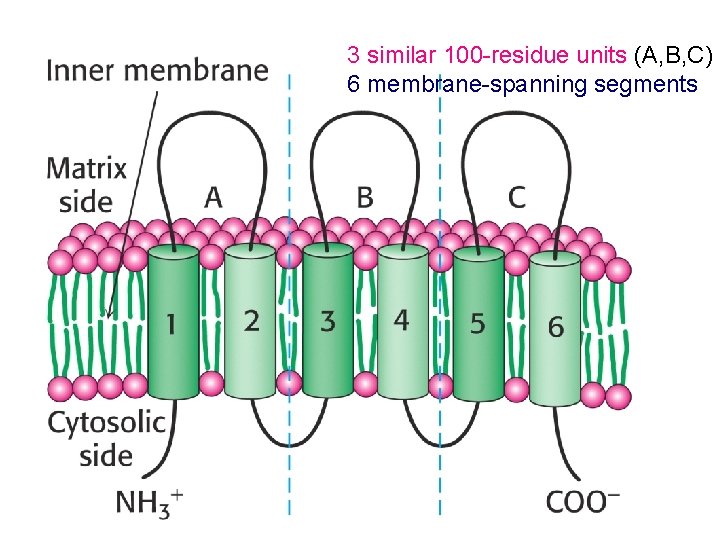
3 similar 100 -residue units (A, B, C) 6 membrane-spanning segments




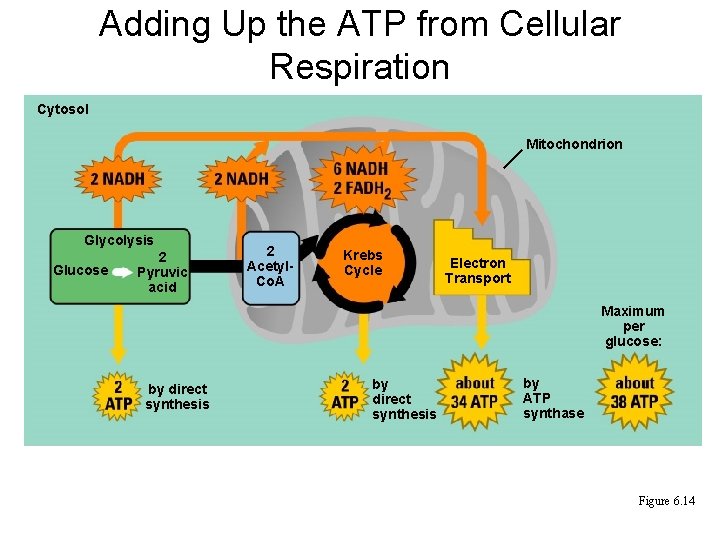
Adding Up the ATP from Cellular Respiration Cytosol Mitochondrion Glycolysis Glucose 2 Pyruvic acid 2 Acetyl. Co. A Krebs Cycle Electron Transport Maximum per glucose: by direct synthesis by ATP synthase Figure 6. 14

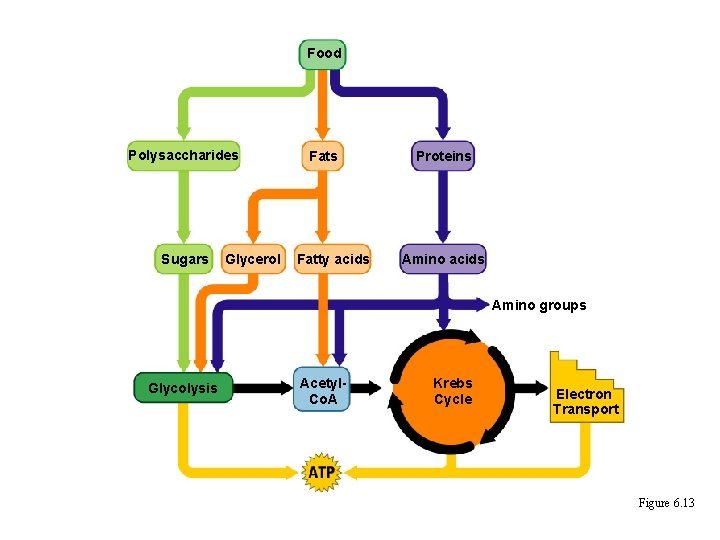
Food Polysaccharides Sugars Glycerol Fats Fatty acids Proteins Amino acids Amino groups Glycolysis Acetyl. Co. A Krebs Cycle Electron Transport Figure 6. 13