Assigning Transmembrane Segments to Helices in IntermediateResolution Structures

Assigning Transmembrane Segments to Helices in Intermediate-Resolution Structures Angela Enosh Sarel J. Fleishman Nir Ben-Tal & Dan Halperin Adapted from a presentation made by Angela Enosh

Lecture Outline n n Background The assignment problem The algorithm Validation

TM proteins form bundles n n helix Transmembrane (TM) proteins cross membrane planes Constitute approximately 50% of contemporary drug targets Helices typically cross the membrane Loops are typically located on the external/internal side of the membrane, connecting consecutive helices Figure 1: 3 D structure of Bacteriorhodopsin
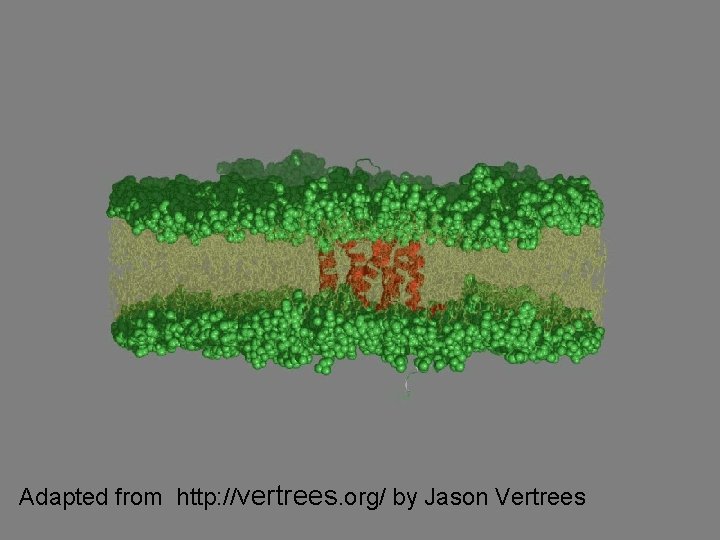
Adapted from http: //vertrees. org/ by Jason Vertrees
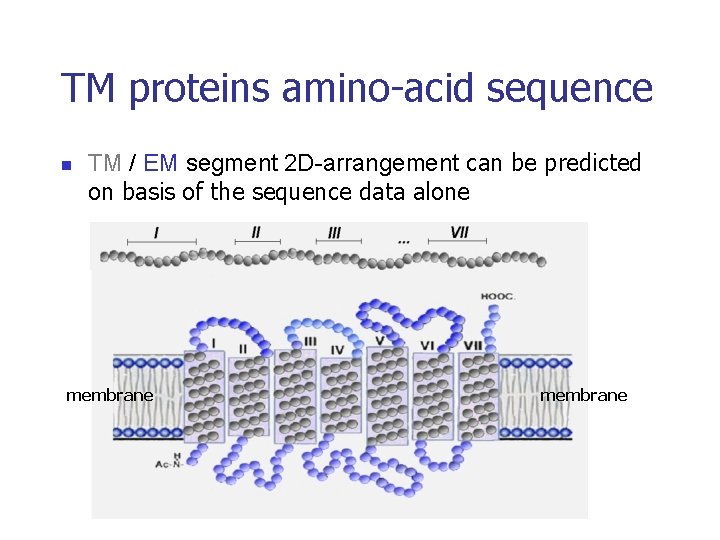
TM proteins amino-acid sequence n TM / EM segment 2 D-arrangement can be predicted on basis of the sequence data alone membrane
TM protein 3 D structure n n Technical problems hamper TM protein structure determination Only 30 distinct folds have been solved using high resolution methods such as X-ray crystallography
Cryo-electron microscopy (Cryo-EM) n n Determines protein structure with low resolution ( >4Å) Individual amino-acids cannot be identified Supplies the locations of the helices Exact structure is left ambiguous

Cryo-electron microscopy (cryo-EM) ** Bovine rhodopsin; adapted from Krebs et al. (2003) J. Biol. Chem. 278, 50217.

Problem description Input and Target n n Position, orientation and azimuth of helices with respect to the membrane planes Partitioning of the sequence into TM segments (helices) and extra membrane segments (loops) Target: Find correspondence between the TM helix-segments and the cryo-EM helices Attempt to reduce the number of possible assignments
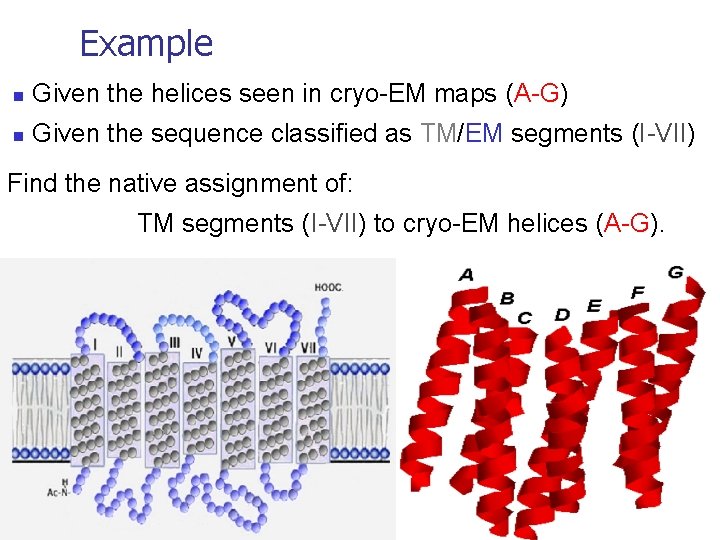
Example n Given the helices seen in cryo-EM maps (A-G) n Given the sequence classified as TM/EM segments (I-VII) Find the native assignment of: TM segments (I-VII) to cryo-EM helices (A-G).
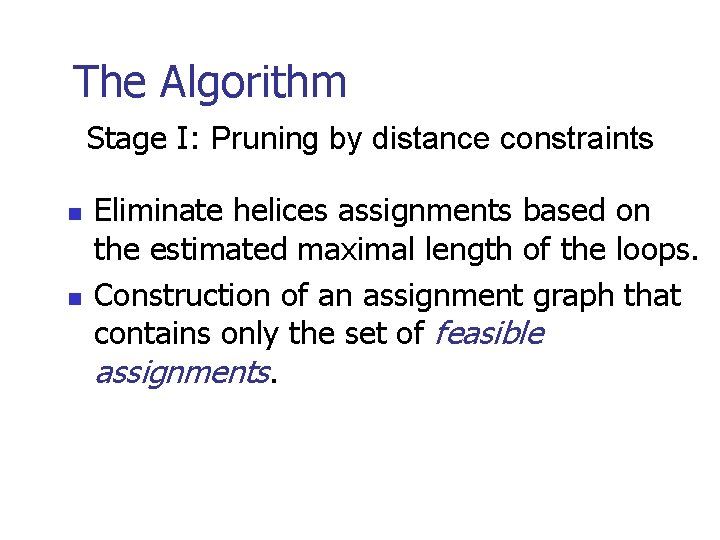
The Algorithm Stage I: Pruning by distance constraints n n Eliminate helices assignments based on the estimated maximal length of the loops. Construction of an assignment graph that contains only the set of feasible assignments.
The Algorithm Stage II: Ranking the feasible assignments n n Use known protein structures taken from the Protein Data Bank (PDB) Score each assignment based on the capability of loops to connect pairs of helices in 3 D.
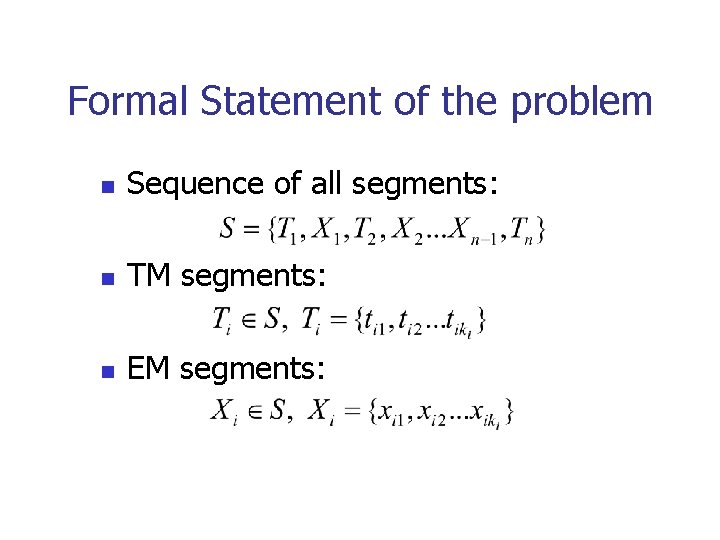
Formal Statement of the problem n Sequence of all segments: n TM segments: n EM segments:
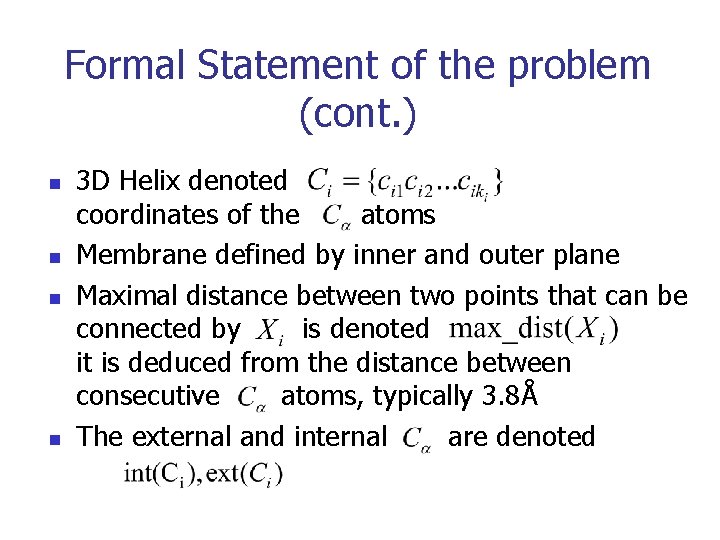
Formal Statement of the problem (cont. ) n n 3 D Helix denoted coordinates of the atoms Membrane defined by inner and outer plane Maximal distance between two points that can be connected by is denoted it is deduced from the distance between consecutive atoms, typically 3. 8Å The external and internal are denoted

Formal Goals n n Find all feasible assignments of ‘s and ‘s An assignment is a permutation where is assigned to Attribute a score to each assignment based on the compatibility with locations of the helices Remark: N-Termini and C-Termini can be deduced experimentally
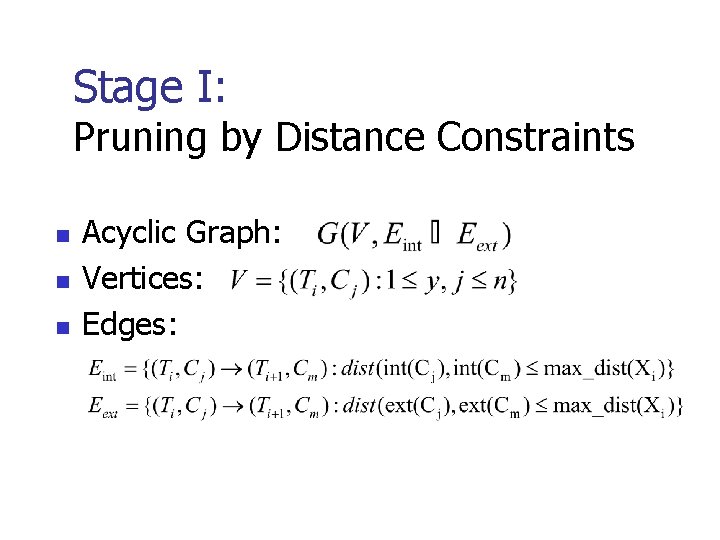
Stage I: Pruning by Distance Constraints n n n Acyclic Graph: Vertices: Edges:
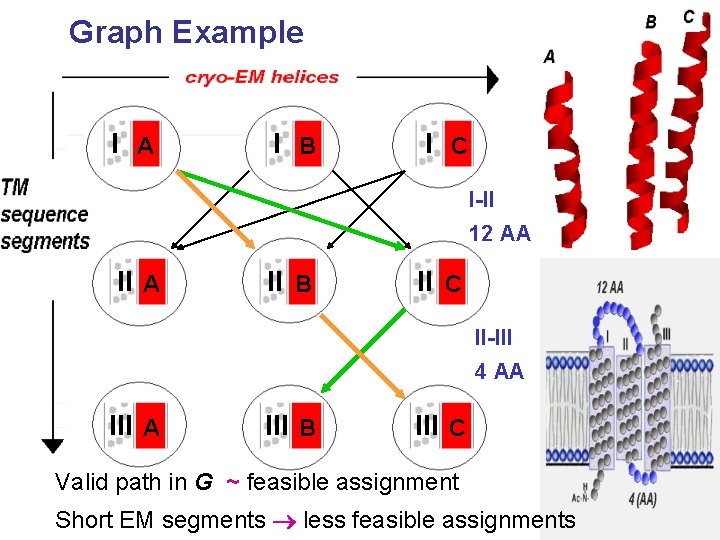
Graph Example I A I B I C C I-II 12 AA II A B II B C II-III 4 AA A III BB III C C Valid path in G ~ feasible assignment Short EM segments less feasible assignments
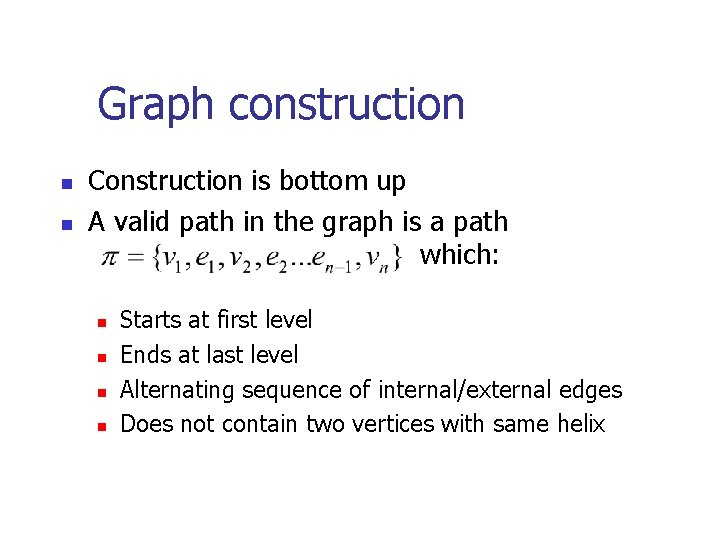
Graph construction n n Construction is bottom up A valid path in the graph is a path which: n n Starts at first level Ends at last level Alternating sequence of internal/external edges Does not contain two vertices with same helix

Stage II: Ranking Feasible Assignments n n n A score is assigned to each feasible assignment stored in G For each we defines the feasibility of connecting two helices in 3 D-space by
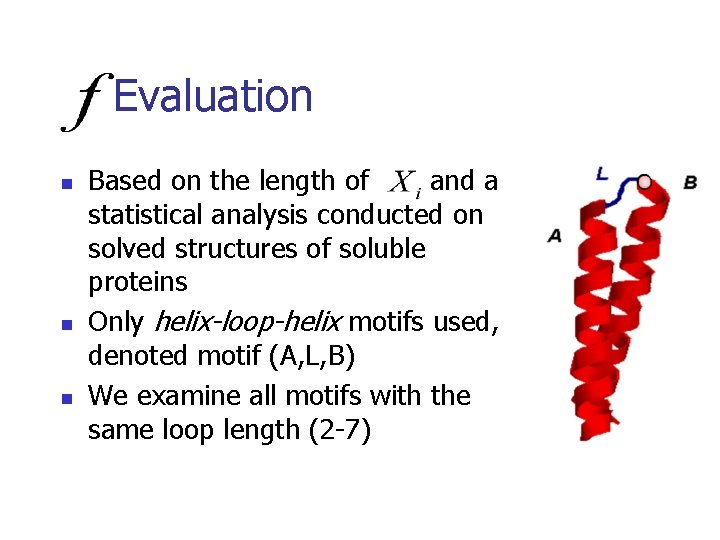
Evaluation n Based on the length of and a statistical analysis conducted on solved structures of soluble proteins Only helix-loop-helix motifs used, denoted motif (A, L, B) We examine all motifs with the same loop length (2 -7)
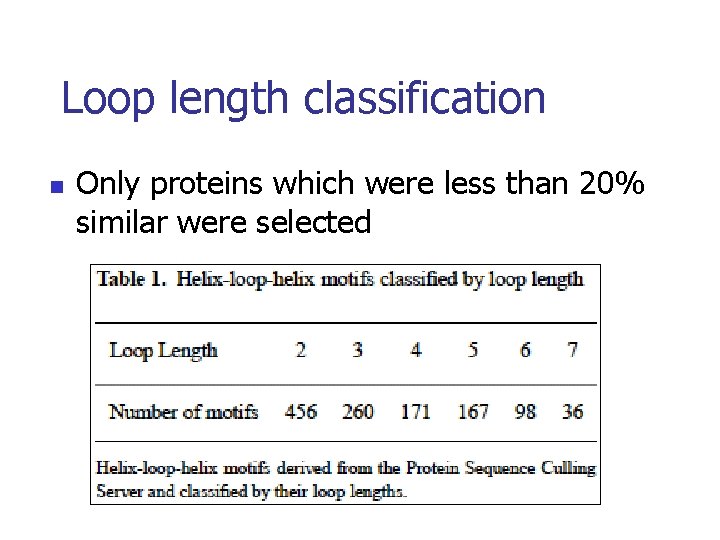
Loop length classification n Only proteins which were less than 20% similar were selected
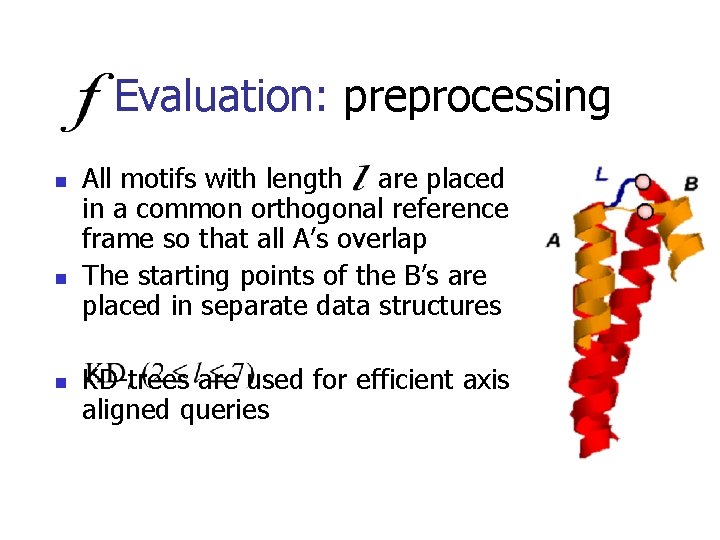
Evaluation: preprocessing n n n All motifs with length are placed in a common orthogonal reference frame so that all A’s overlap The starting points of the B’s are placed in separate data structures KD-trees are used for efficient axis aligned queries
Distribution of end points of short loops n n n Kinematics considerations allow a reachable space limited only by the length of the loop Example: loop length of 4 results in 8 degrees of freedom In reality the end points tend to be highly nonuniform Highly significant with loops of length two to five Still noticeable in loops of lengths up to seven
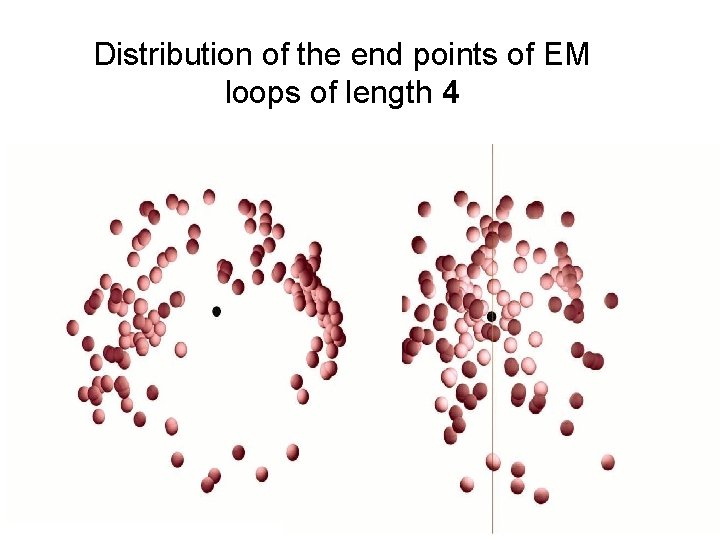
Distribution of the end points of EM loops of length 4
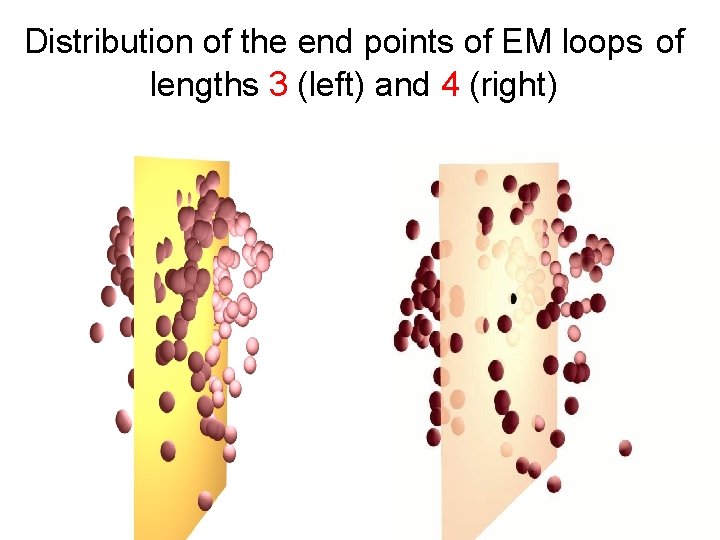
Distribution of the end points of EM loops of lengths 3 (left) and 4 (right)

Evaluation: scoring n The 2 helices are placed in the same reference frame Q is a cube around the start of B with a side size of Å We define a colony function n the score depends on: n n number of neighboring points in the vicinity of q distances between these neighboring points and q

Evaluation n The score of the assignment is the total score of its extra membrane segments Define a weight for each edge For each path we define to be:

Validation n n 19 TM proteins with a known high resolution structure were tested Two distinct cases: • • Accurate data Noisy data regarding the locations and orientations of the helices
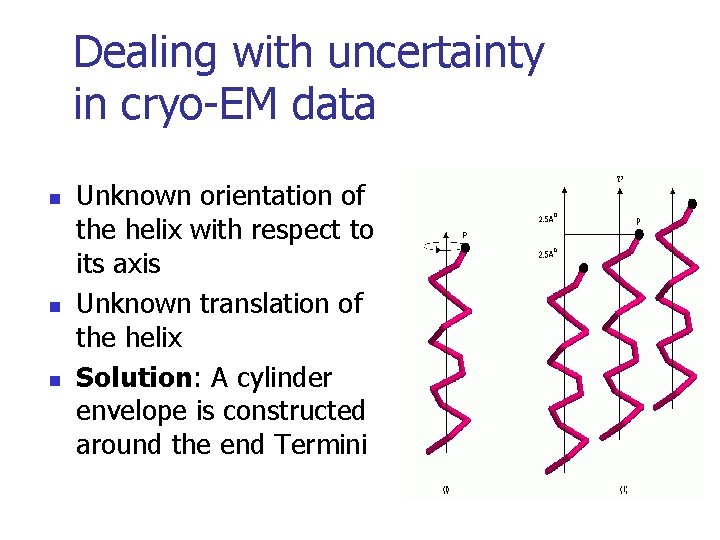
Dealing with uncertainty in cryo-EM data n n n Unknown orientation of the helix with respect to its axis Unknown translation of the helix Solution: A cylinder envelope is constructed around the end Termini
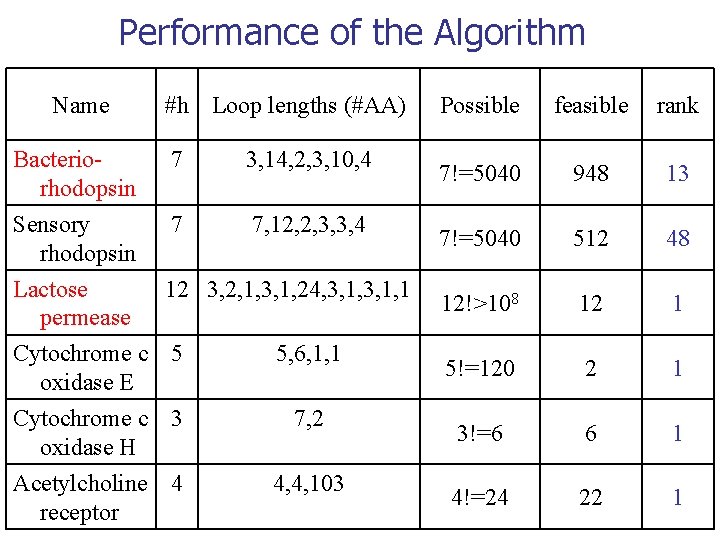
Performance of the Algorithm Name Bacteriorhodopsin #h Loop lengths (#AA) 7 3, 14, 2, 3, 10, 4 Sensory 7 7, 12, 2, 3, 3, 4 rhodopsin Lactose 12 3, 2, 1, 3, 1, 24, 3, 1, 1 permease Cytochrome c 5 5, 6, 1, 1 oxidase E Cytochrome c 3 oxidase H Acetylcholine 4 receptor 7, 2 4, 4, 103 Possible feasible rank 7!=5040 948 13 7!=5040 512 48 12!>108 12 1 5!=120 2 1 3!=6 6 1 4!=24 22 1

Summary n Provides more than a single assignment n The complexity of the problem scales with the number of amino-acids in the extra-membrane segments – not with the number of TM helices

Questions
- Slides: 32