Artificial Intelligence Project 3 Diagnosis Using Bayesian Networks

Artificial Intelligence Project #3 : Diagnosis Using Bayesian Networks May 19, 2005 (c) 2005 SNU CSE Biointelligence Lab
Goals of the Project l Analysis of the influence of network size and data size on structural learning of Bayesian networks ¨ ¨ l Six Bayesian networks of various sizes are given. Generate data examples from each Bayesian network. Learn Bayesian network structures from the generated data. Analyze the learned results according to the network size and the data size. Classification using Bayesian networks ¨ A microarray dataset consisting of two classes of samples is given. ¨ Learn Bayesian network classifiers from the dataset. ¨ Compare the classification accuracy of Bayesian network classifiers with that of other classifiers such as neural networks. (c) 2005 SNU CSE Biointelligence Lab 2

Given Bayesian Networks Randomly generated l Network structure: scale-free and modular l # of variables: 10, 30, and 45 l All variables are binary l l Network file format: *. dsc for MSBNX (http: //research. microsoft. com/adapt/MSBNx/) (c) 2005 SNU CSE Biointelligence Lab 3
Example Bayesian Network Structure I (c) 2005 SNU CSE Biointelligence Lab 4
Example Bayesian Network Structure II (c) 2005 SNU CSE Biointelligence Lab 5
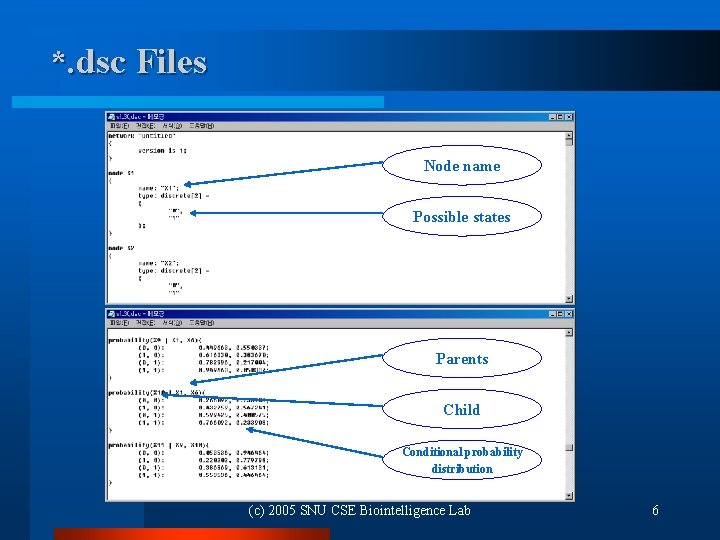
*. dsc Files Node name Possible states Parents Child Conditional probability distribution (c) 2005 SNU CSE Biointelligence Lab 6
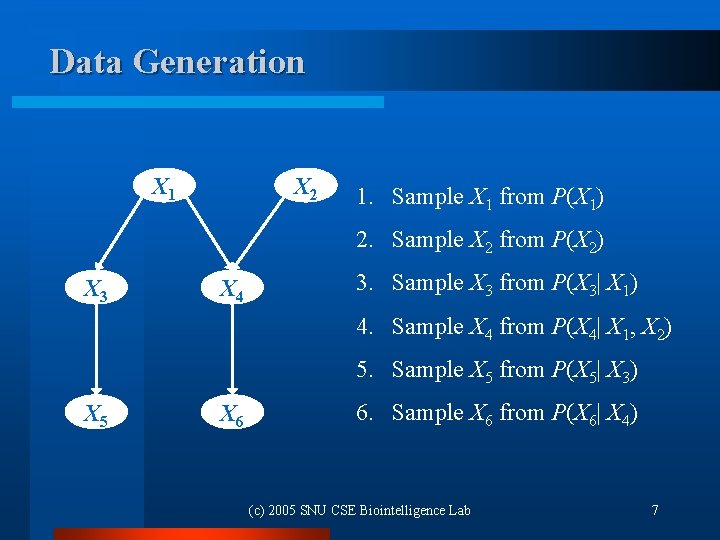
Data Generation X 1 X 2 1. Sample X 1 from P(X 1) 2. Sample X 2 from P(X 2) X 3 X 4 3. Sample X 3 from P(X 3| X 1) 4. Sample X 4 from P(X 4| X 1, X 2) 5. Sample X 5 from P(X 5| X 3) X 5 X 6 6. Sample X 6 from P(X 6| X 4) (c) 2005 SNU CSE Biointelligence Lab 7
![Data Generation Tool l data_generator ¨ Usage: data_generator [network file style] [# of nodes] Data Generation Tool l data_generator ¨ Usage: data_generator [network file style] [# of nodes]](http://slidetodoc.com/presentation_image_h/00608063cf55188145096faf54aee8bc/image-8.jpg)
Data Generation Tool l data_generator ¨ Usage: data_generator [network file style] [# of nodes] [# of data samples] [input file] [output file]. . . (c) 2005 SNU CSE Biointelligence Lab 8
Structural Learning of Bayesian Networks l Using WEKA software (http: //www. cs. waikato. ac. nz/ml/weka/) (c) 2005 SNU CSE Biointelligence Lab 9
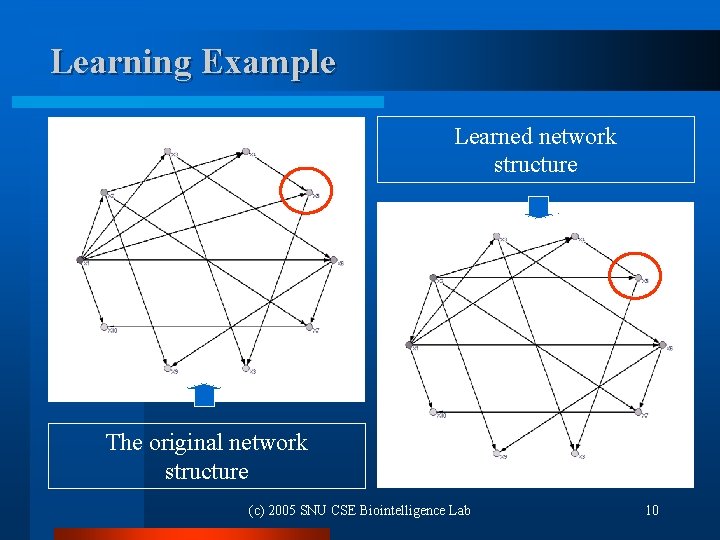
Learning Example Learned network structure The original network structure (c) 2005 SNU CSE Biointelligence Lab 10

Materials for the First One l Given ¨ Bayesian networks < sf_10. dsc, sf_30. dsc, sf_45. dsc, md_10. dsc, md_30. dsc, md_45. dsc ¨ Data generation tool < data_generator. exe l [for Windows], data_generator [for Linux] Downloadable ¨ MSBNX (http: //research. microsoft. com/adapt/MSBNx/) ¨ WEKA (http: //www. cs. waikato. ac. nz/ml/weka/) l You should write your own code for comparing Bayesian network structures. (c) 2005 SNU CSE Biointelligence Lab 11

Study l Treatment-specific changes in gene expression discriminate in vivo drug response in human leukemia cells, MH Cheok et al. , Nature Genetics 35, 2003. 60 leukemia patients Bone marrow samples Affymetrix Gene. Chip arrays (c) 2005 SNU CSE Biointelligence Lab Gene expression data 12

Gene Expression Data l # of data examples ¨ 120 (60: before treatment, 60: after treatment) l # of genes measured ¨ 12600 (Affymetrix HG-U 95 A array) l Task ¨ Classification between “before treatment” and “after treatment” based on gene expression pattern (c) 2005 SNU CSE Biointelligence Lab 13
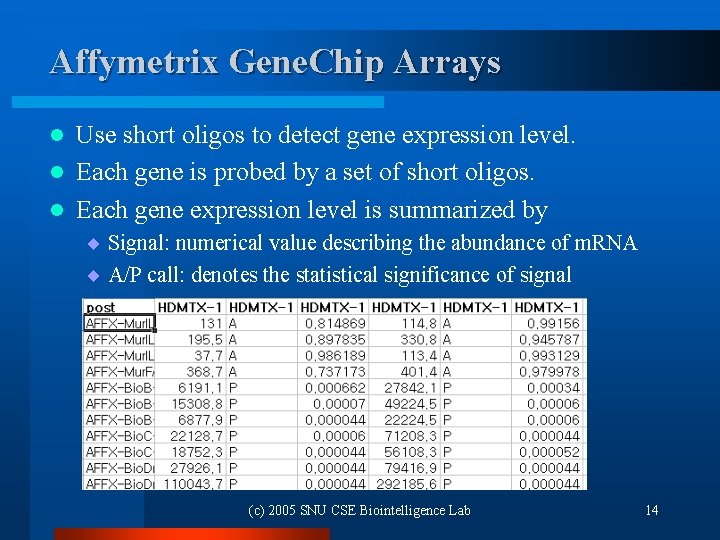
Affymetrix Gene. Chip Arrays Use short oligos to detect gene expression level. l Each gene is probed by a set of short oligos. l Each gene expression level is summarized by l ¨ Signal: numerical value describing the abundance of m. RNA ¨ A/P call: denotes the statistical significance of signal (c) 2005 SNU CSE Biointelligence Lab 14

Preprocessing l Remove the genes having more than 60 ‘A’ calls ¨ # of genes: 12600 3190 l Discretization of gene expression level ¨ Criterion: median gene expression value of each sample ¨ 0 (low) and 1 (high) (c) 2005 SNU CSE Biointelligence Lab 15
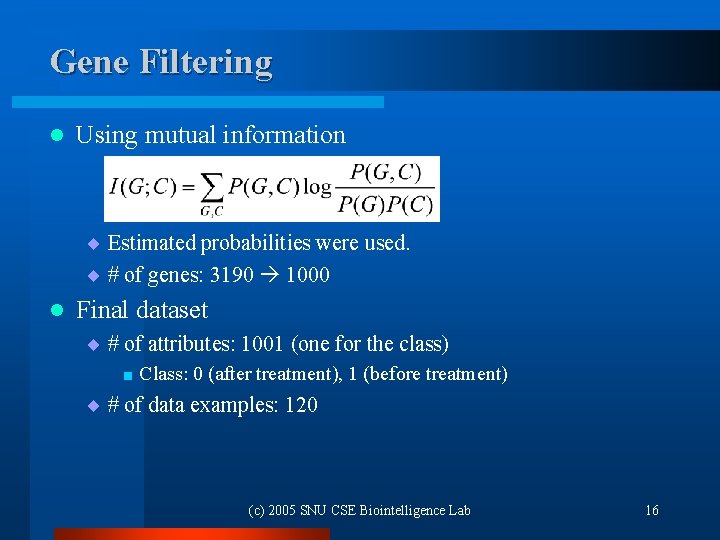
Gene Filtering l Using mutual information ¨ Estimated probabilities were used. ¨ # of genes: 3190 1000 l Final dataset ¨ # of attributes: 1001 (one for the class) < Class: 0 (after treatment), 1 (before treatment) ¨ # of data examples: 120 (c) 2005 SNU CSE Biointelligence Lab 16
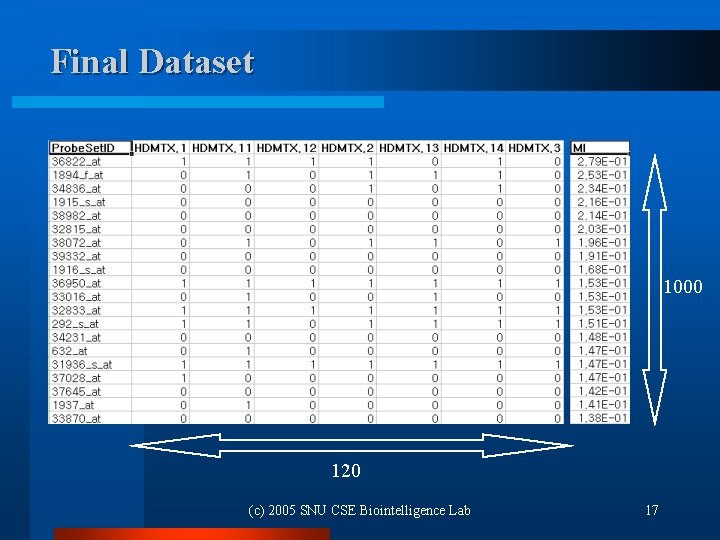
Final Dataset 1000 120 (c) 2005 SNU CSE Biointelligence Lab 17

Materials for the Second One l Given ¨ Preprocessed microarray data file: data 2. txt l Downloadable ¨ WEKA (http: //www. cs. waikato. ac. nz/ml/weka/) (c) 2005 SNU CSE Biointelligence Lab 18
Due: June 16, 2005 l Analysis of the influence of network size and data size on structural learning of Bayesian networks ¨ ¨ l Six Bayesian networks of various sizes are given. Generate data examples from each Bayesian network. Learn Bayesian network structures from the generated data. Analyze the learned results according to the network size and the data size. Classification using Bayesian networks ¨ A microarray dataset consisting of two classes of samples is given. ¨ Learn Bayesian network classifiers from the dataset. ¨ Compare the classification accuracy of Bayesian network classifiers with that of other classifiers such as neural networks. (c) 2005 SNU CSE Biointelligence Lab 19
- Slides: 19