Artificial Immune Systems Andrew Watkins Why the Immune














- Slides: 47

Artificial Immune Systems Andrew Watkins

Why the Immune System? • Recognition – Anomaly detection – Noise tolerance • • Robustness Feature extraction Diversity Reinforcement learning Memory Distributed Multi-layered Adaptive

Definition AIS are adaptive systems inspired by theoretical immunology and observed immune functions, principles and models, which are applied to complex problem domains (de Castro and Timmis)

Some History • Developed from the field of theoretical immunology in the mid 1980’s. – Suggested we ‘might look’ at the IS • 1990 – Bersini first use of immune algos to solve problems • Forrest et al – Computer Security mid 1990’s • Hunt et al, mid 1990’s – Machine learning

How does it work?

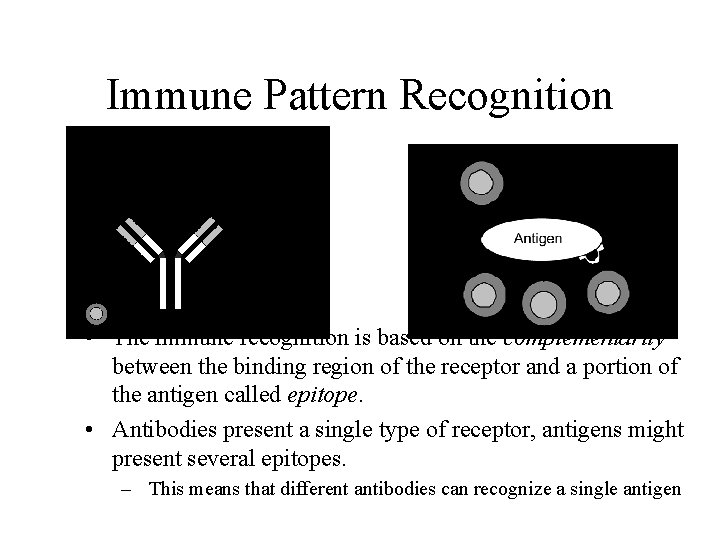
Immune Pattern Recognition • The immune recognition is based on the complementarity between the binding region of the receptor and a portion of the antigen called epitope. • Antibodies present a single type of receptor, antigens might present several epitopes. – This means that different antibodies can recognize a single antigen

Immune Responses

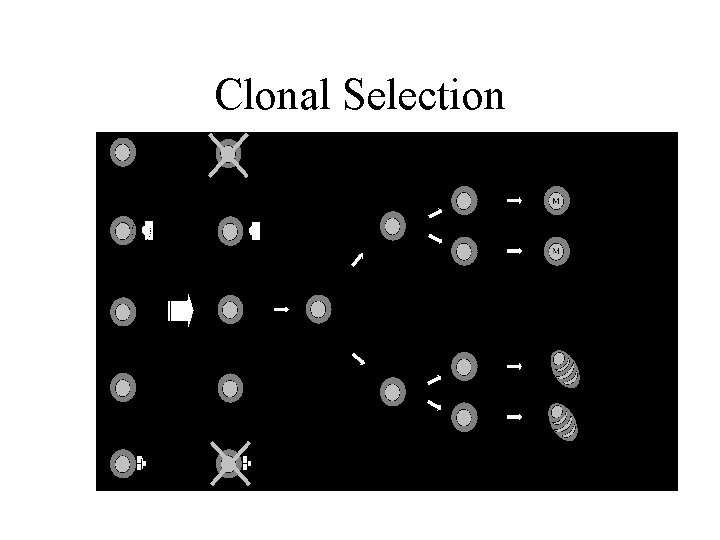
Clonal Selection

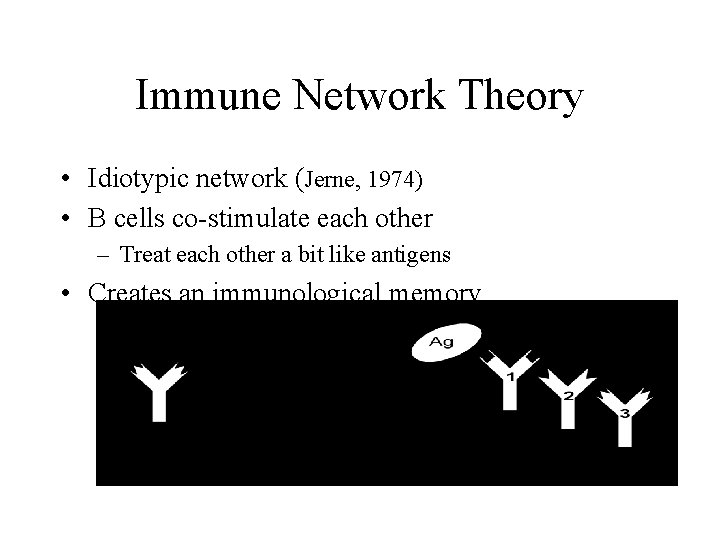
Immune Network Theory • Idiotypic network (Jerne, 1974) • B cells co-stimulate each other – Treat each other a bit like antigens • Creates an immunological memory

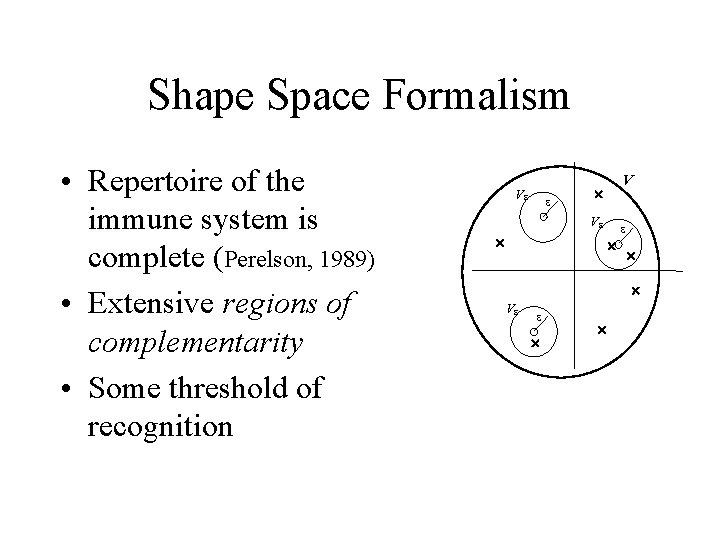
Shape Space Formalism • Repertoire of the immune system is complete (Perelson, 1989) • Extensive regions of complementarity • Some threshold of recognition Ve e V ´ Ve ´ ´ Ve e ´ ´

Self/Non-Self Recognition • Immune system needs to be able to differentiate between self and non-self cells • Antigenic encounters may result in cell death, therefore – Some kind of positive selection – Some element of negative selection

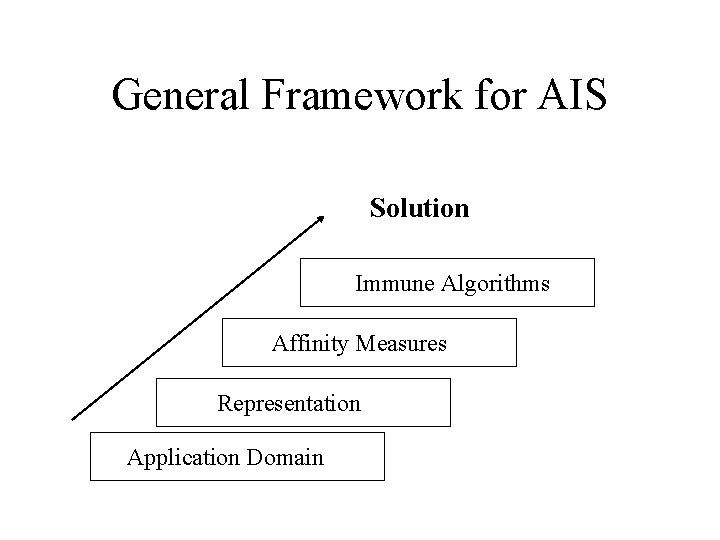
General Framework for AIS Solution Immune Algorithms Affinity Measures Representation Application Domain

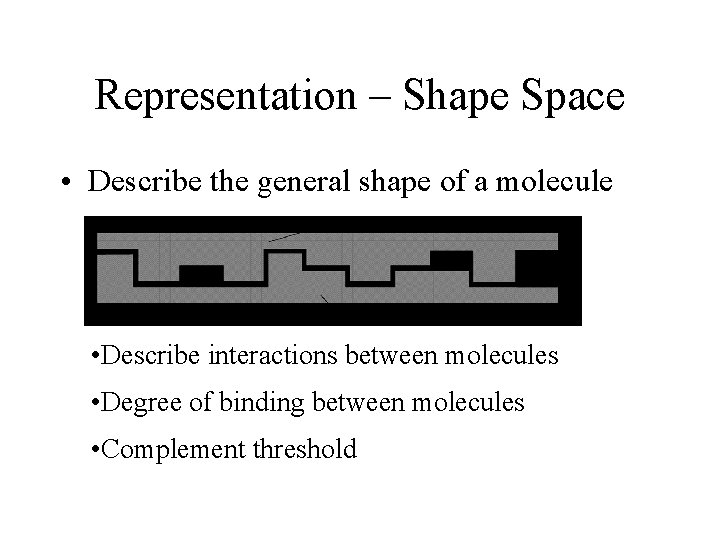
Representation – Shape Space • Describe the general shape of a molecule • Describe interactions between molecules • Degree of binding between molecules • Complement threshold

Define their Interaction • Define the term Affinity • Affinity is related to distance – Euclidian • Other distance measures such as Hamming, Manhattan etc. • Affinity Threshold

Basic Immune Models and Algorithms • • • Bone Marrow Models Negative Selection Algorithms Clonal Selection Algorithm Somatic Hypermutation Immune Network Models

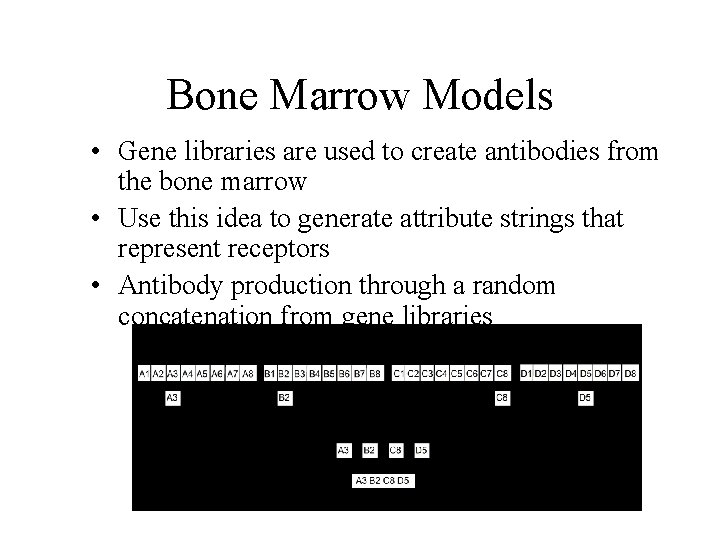
Bone Marrow Models • Gene libraries are used to create antibodies from the bone marrow • Use this idea to generate attribute strings that represent receptors • Antibody production through a random concatenation from gene libraries

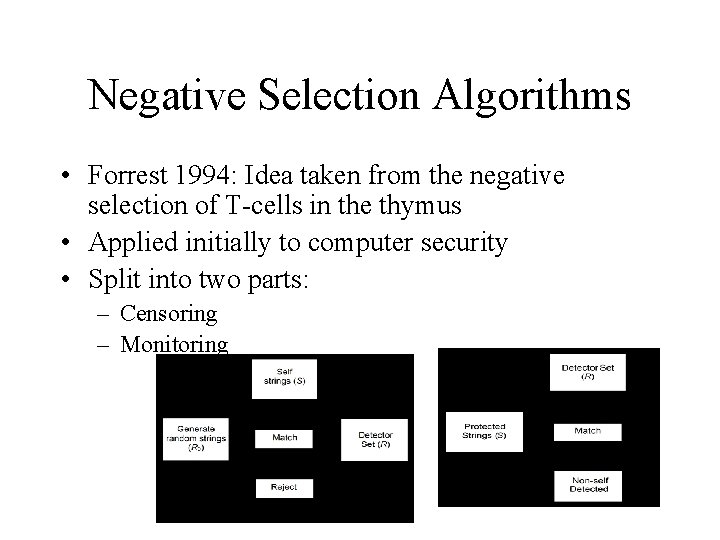
Negative Selection Algorithms • Forrest 1994: Idea taken from the negative selection of T-cells in the thymus • Applied initially to computer security • Split into two parts: – Censoring – Monitoring

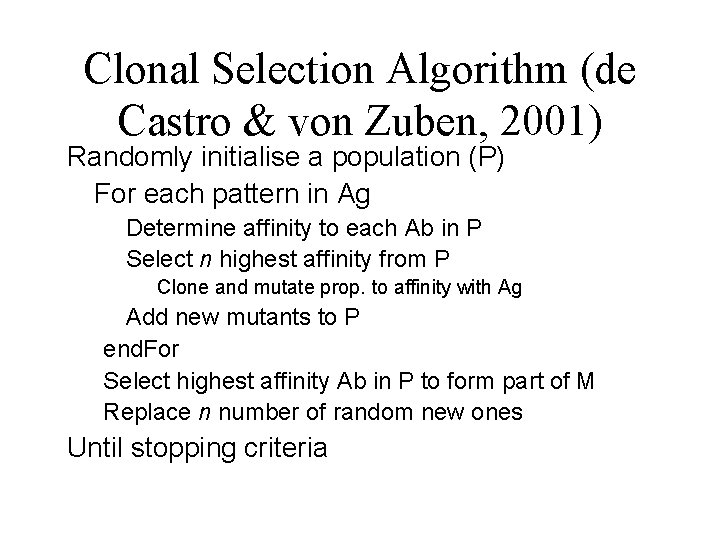
Clonal Selection Algorithm (de Castro & von Zuben, 2001) Randomly initialise a population (P) For each pattern in Ag Determine affinity to each Ab in P Select n highest affinity from P Clone and mutate prop. to affinity with Ag Add new mutants to P end. For Select highest affinity Ab in P to form part of M Replace n number of random new ones Until stopping criteria

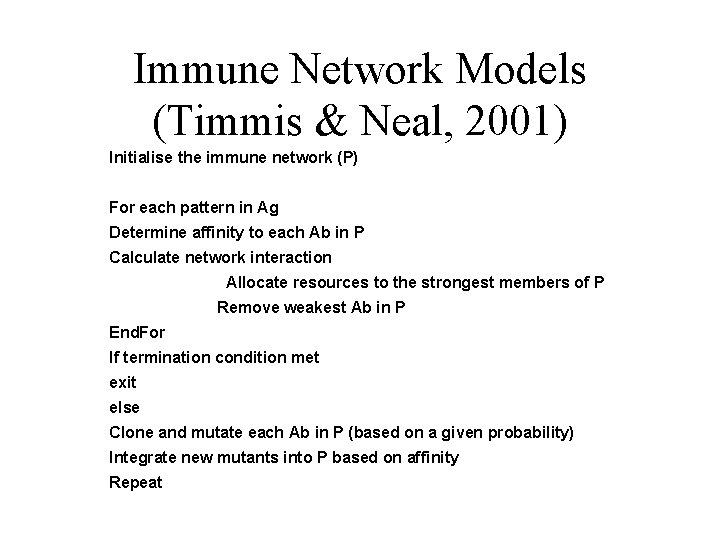
Immune Network Models (Timmis & Neal, 2001) Initialise the immune network (P) For each pattern in Ag Determine affinity to each Ab in P Calculate network interaction Allocate resources to the strongest members of P Remove weakest Ab in P End. For If termination condition met exit else Clone and mutate each Ab in P (based on a given probability) Integrate new mutants into P based on affinity Repeat

Somatic Hypermutation • Mutation rate in proportion to affinity • Very controlled mutation in the natural immune system • The greater the antibody affinity the smaller its mutation rate • Classic trade-off between exploration and exploitation

How do AIS Compare? • Basic Components: – AIS B-cell in shape space (e. g. attribute strings) • Stimulation level – ANN Neuron • Activation function – GA chromosome • fitness

Comparing • Structure (Architecture) – AIS and GA fixed or variable sized populations, not connected in population based AIS – ANN and AIS • Do have network based AIS • ANN typically fixed structure (not always) • Learning takes place in weights in ANN

Comparing • Memory – AIS in B-cells • Network models in connections – ANN In weights of connections – GA individual chromosome

Comparing • • • Adaptation Dynamics Metadynamics Interactions Generalisation capabilities Etc. many more.

Where are they used? • • • Dependable systems Scheduling Robotics Security Anomaly detection Learning systems

Artificial Immune Recognition System (AIRS): An Immune-Inspired Supervised Learning Algorithm

AIRS: Immune Principles Employed • Clonal Selection • Based initially on immune networks, though found this did not work • Somatic hypermutation – Eventually • Recognition regions within shape space • Antibody/antigen binding

AIRS: Mapping from IS to AIS • Antibody • Recognition Ball (RB) • Antigens • Immune Memory Feature Vector Combination of feature vector and vector class Training Data Memory cells—set of mutated Artificial RBs

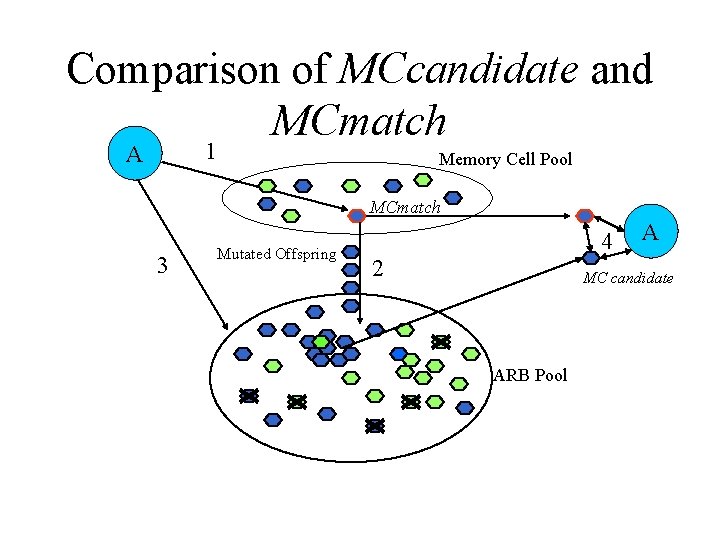
Classification • Stimulation of an ARB is based not only on its affinity to an antigen but also on its class when compared to the class of an antigen • Allocation of resources to the ARBs also takes into account the ARBs’ classifications when compared to the class of the antigen • Memory cell hyper-mutation and replacement is based primarily on classification and secondarily on affinity

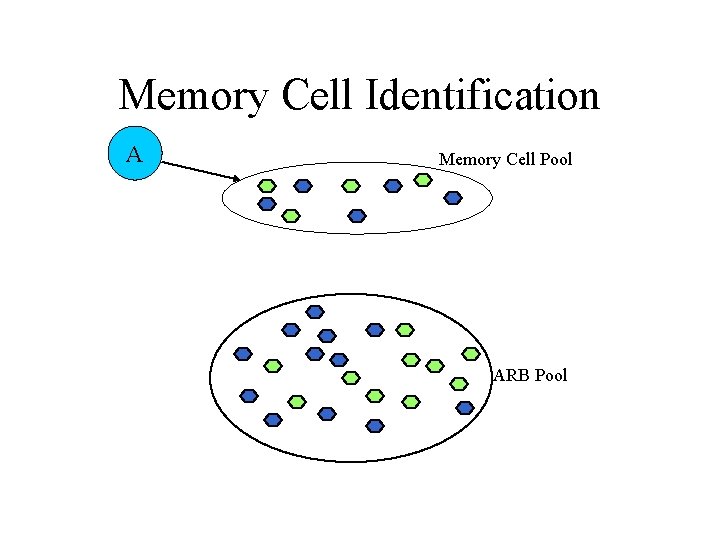
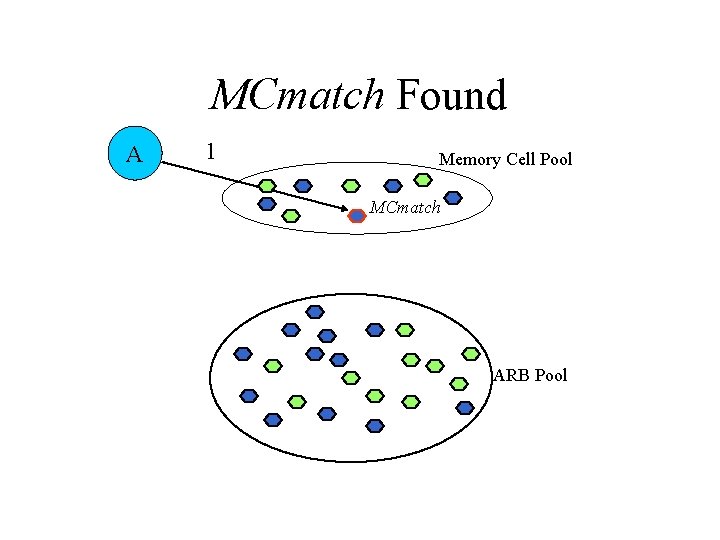
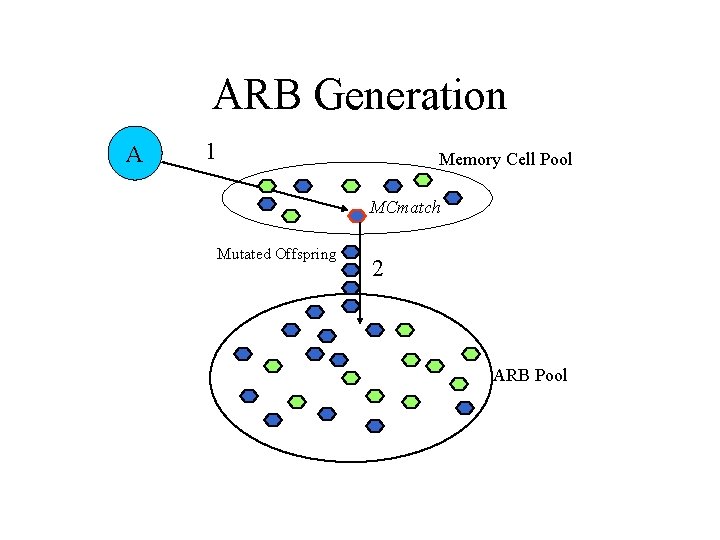
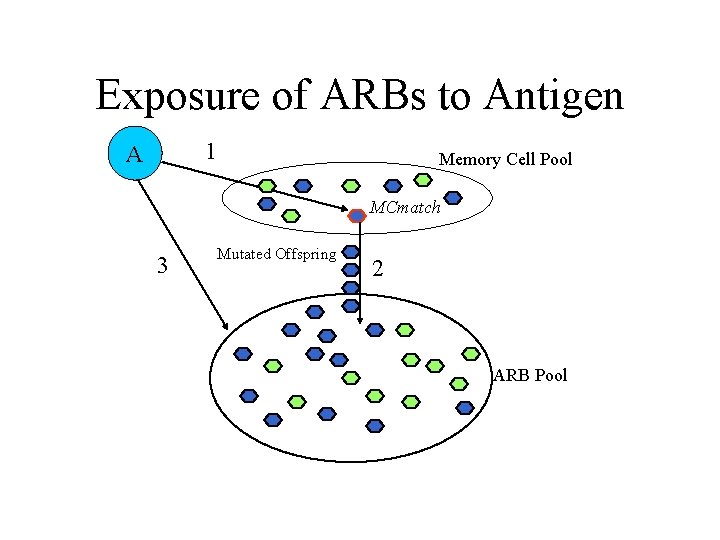
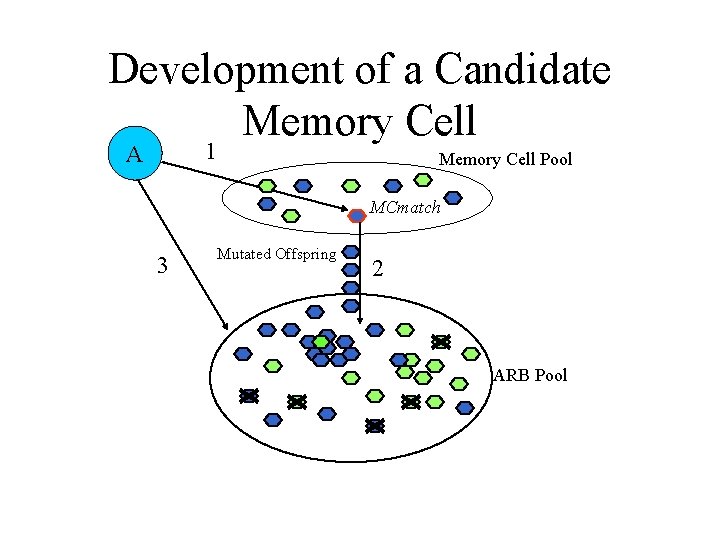
AIRS Algorithm • Data normalization and initialization • Memory cell identification and ARB generation • Competition for resources in the development of a candidate memory cell • Potential introduction of the candidate memory cell into the set of established memory cells

Memory Cell Identification A Memory Cell Pool ARB Pool

MCmatch Found A 1 Memory Cell Pool MCmatch ARB Pool

ARB Generation A 1 Memory Cell Pool MCmatch Mutated Offspring 2 ARB Pool

Exposure of ARBs to Antigen 1 A Memory Cell Pool MCmatch 3 Mutated Offspring 2 ARB Pool

Development of a Candidate Memory Cell 1 A Memory Cell Pool MCmatch 3 Mutated Offspring 2 ARB Pool

Comparison of MCcandidate and MCmatch 1 A Memory Cell Pool MCmatch 3 Mutated Offspring 4 2 A MC candidate ARB Pool

Memory Cell Introduction 1 A Memory Cell Pool MCmatch 3 Mutated Offspring 2 4 5 A MCcandidate ARB Pool

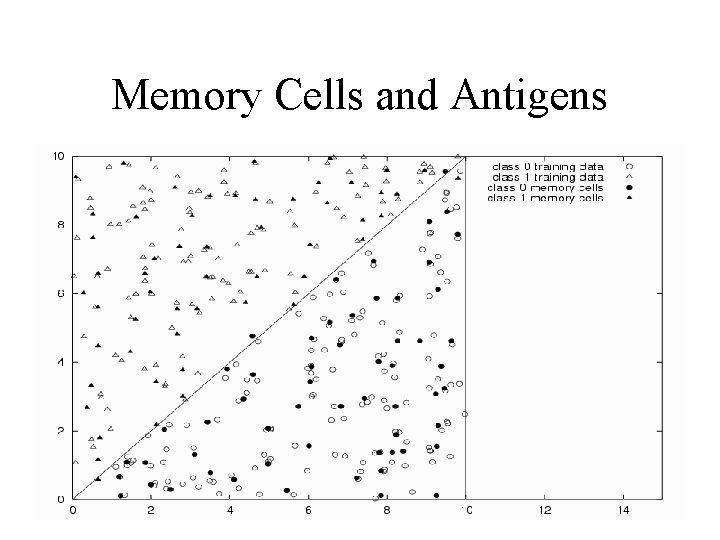
Memory Cells and Antigens

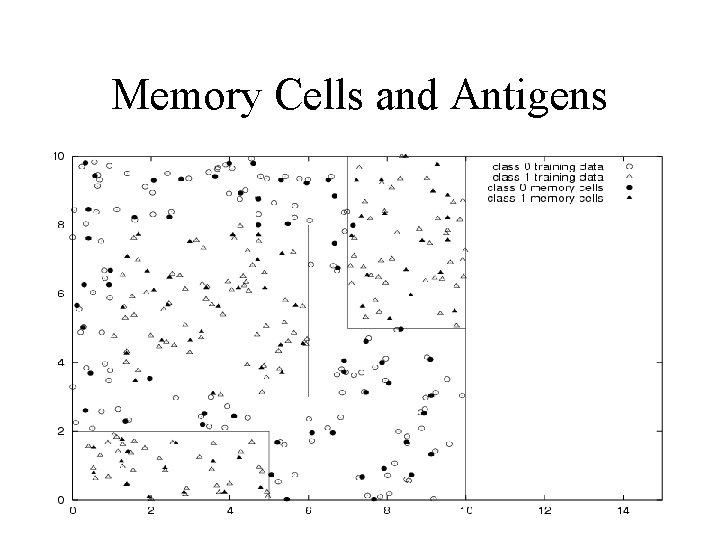
Memory Cells and Antigens

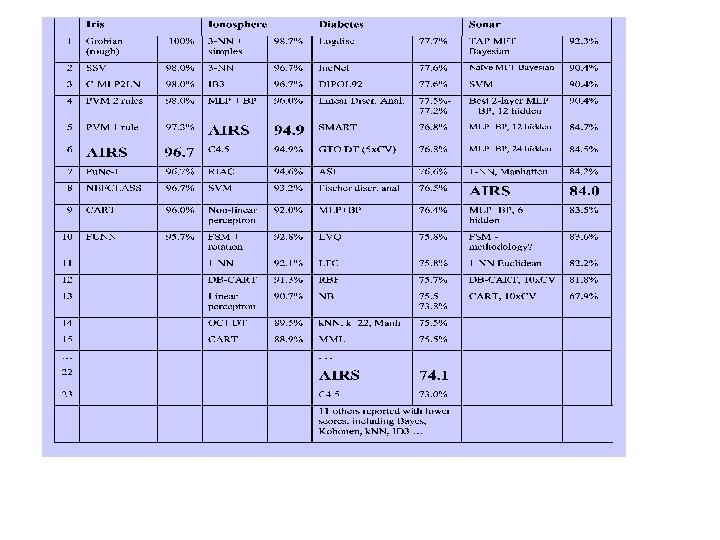
AIRS: Performance Evaluation Fisher’s Iris Data Set Pima Indians Diabetes Data Set Ionosphere Data Set Sonar Data Set


AIRS: Observations • ARB Pool formulation was over complicated – Crude visualization – Memory only needs to be maintained in the Memory Cell Pool • Mutation Routine – Difference in Quality – Some redundancy

AIRS: Revisions • Memory Cell Evolution – Only Memory Cell Pool has different classes – ARB Pool only concerned with evolving memory cells • Somatic Hypermutation – Cell’s stimulation value indicates range of mutation possibilities – No longer need to mutate class

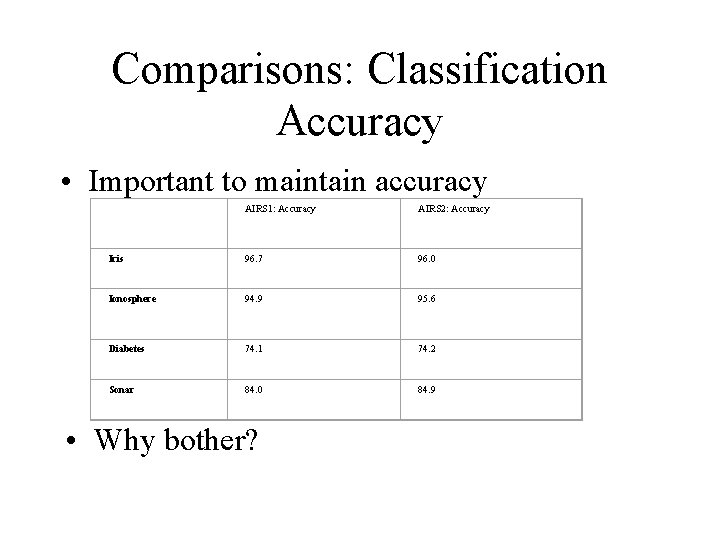
Comparisons: Classification Accuracy • Important to maintain accuracy AIRS 1: Accuracy AIRS 2: Accuracy Iris 96. 7 96. 0 Ionosphere 94. 9 95. 6 Diabetes 74. 1 74. 2 Sonar 84. 0 84. 9 • Why bother?

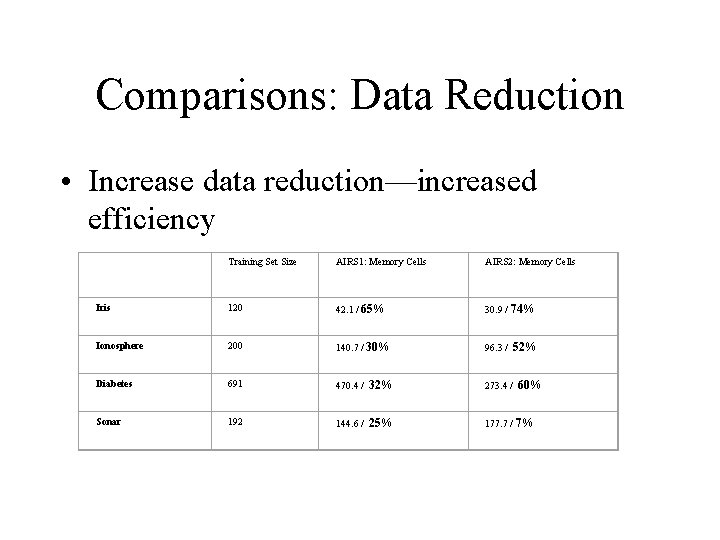
Comparisons: Data Reduction • Increase data reduction—increased efficiency Training Set Size AIRS 1: Memory Cells AIRS 2: Memory Cells Iris 120 42. 1 / 65% 30. 9 / 74% Ionosphere 200 140. 7 / 30% 96. 3 / 52% Diabetes 691 470. 4 / 32% 273. 4 / 60% Sonar 192 144. 6 / 25% 177. 7 / 7%

Features of AIRS • No need to know best architecture to get good results • Default settings within a few percent of the best it can get • User-adjustable parameters optimize performance for a given problem set • Generalization and data reduction

More Information • http: //www. cs. ukc. ac. uk/people/rpg/abw 5 • http: //www. cs. ukc. ac. uk/people/staff/jt 6 • http: //www. cs. ukc. ac. uk/aisbook