Architecture of large projects in bioinformatics ADP Lecture

Architecture of large projects in bioinformatics (ADP) Lecture 02 Łukasz P. Kozłowski lukaskoz@mimuw. edu. pl Warsaw, 2021

Architecture of large projects in bioinformatics (ADP) Lecture 02 https: //www. mimuw. edu. pl/~lukaskoz/teaching/adp/ Łukasz P. Kozłowski lukaskoz@mimuw. edu. pl Warsaw, 2021

People Outline 1. Data formats in bioinformatics 2. Popular software libraries (Bio. Perl, Bio. Python) 3. Most important bioinformatics databases (Uni. Prot, PDB, Ref. Seq, Gen. Bank, ENA, Inter. Pro, etc. ) 4. Software licensing for scientific purposes. Free-software licensing. Patents. 5. Generic model Organism database (GMOD) project assumptions, history and usage 6. Genome browsers, problem description and state of the solutions ADP

People Outline 1. Data formats in bioinformatics 2. Popular software libraries (Bio. Perl, Bio. Python) 3. Most important bioinformatics databases (Uni. Prot, PDB, Ref. Seq, Gen. Bank, ENA, Inter. Pro, etc. ) 4. Software licensing for scientific purposes. Free-software licensing. Patents. 5. Generic model Organism database (GMOD) project assumptions, history and usage 6. Genome browsers, problem description and state of the solutions ADP

Data formats ADP Few words about proteins (interactive part) Protein disorder http: //iimcb. genesilico. pl/metadisorder/protein_disorder_intrinsically_unstructured_proteins_ gallery_images. html Protein modeling (USCF Chimera part)
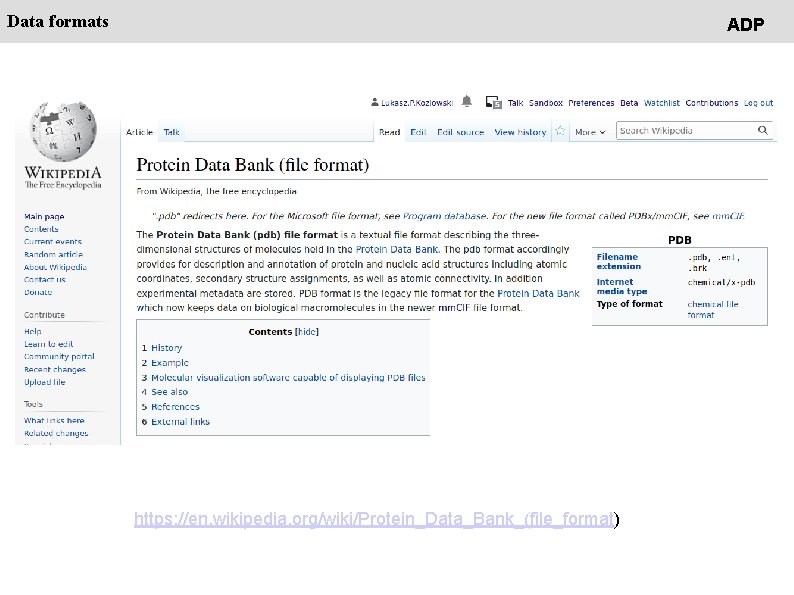
Data formats ADP https: //en. wikipedia. org/wiki/Protein_Data_Bank_(file_format)

People Rules, grading, etc. ADP The essay: mini-review about specific bioinformatics topic Exemplary subjects Review about available software for: – Structural biology (proteins, RNA, drugs) – Phylogenetics – NGS – Chemoinformatics – Data warehouse in bioinformatics (e. g. Biomart) – Genomics (e. g. chip-seq) – Machine learning (clustering, classification, deep learning, etc. ) – Image processing from microscopes/scanners etc. – own suggestions. . . ? You had 1 week to decide/find the subject. Please send your proposition to lukaskoz@mimuw. edu. pl with the email subject ADP 21_essay_Surname_Name

People Popular software libraries Perl → Bio. Perl ADP

People Popular software libraries Perl → Bio. Perl Php → Bio. PHP ADP
People Popular software libraries Perl → Bio. Perl Php → Bio. PHP Java → Bio. Java ADP
People Popular software libraries Perl → Bio. Perl Php → Bio. PHP Java → Bio. Java R →Bioconductor ADP
People Popular software libraries Perl → Bio. Perl Php → Bio. PHP Java → Bio. Java R →Bioconductor Rust → Rust-Bio ADP
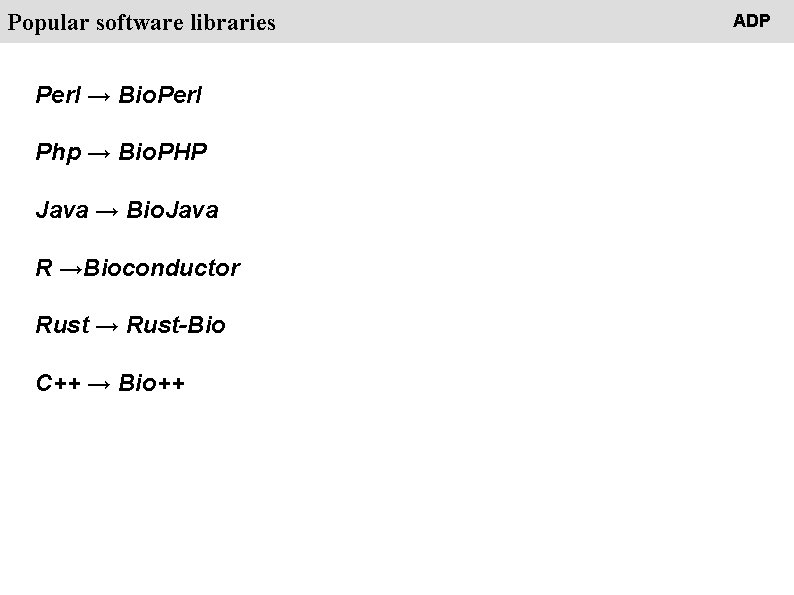
People Popular software libraries Perl → Bio. Perl Php → Bio. PHP Java → Bio. Java R →Bioconductor Rust → Rust-Bio C++ → Bio++ ADP
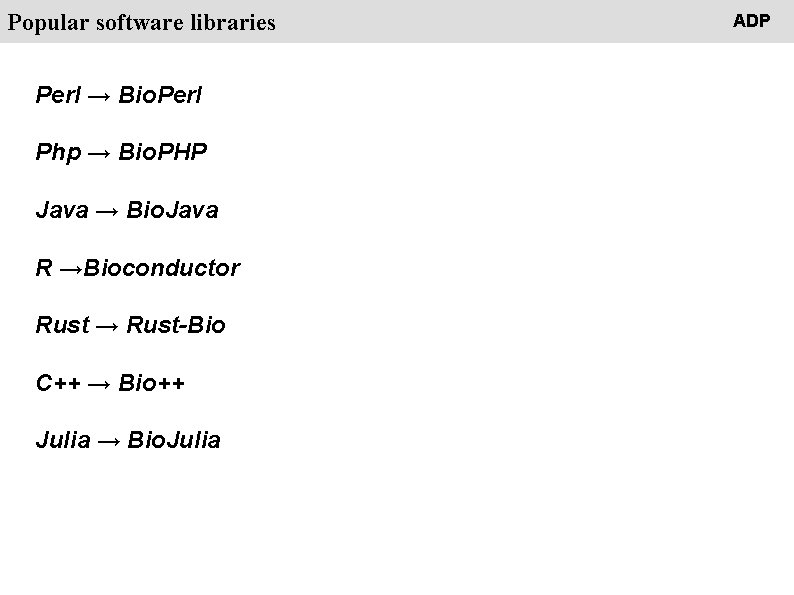
People Popular software libraries Perl → Bio. Perl Php → Bio. PHP Java → Bio. Java R →Bioconductor Rust → Rust-Bio C++ → Bio++ Julia → Bio. Julia ADP
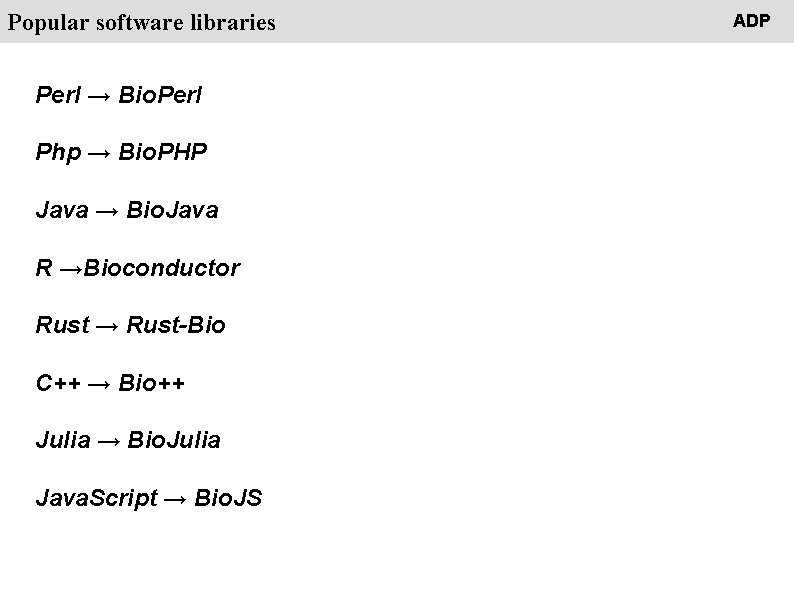
People Popular software libraries Perl → Bio. Perl Php → Bio. PHP Java → Bio. Java R →Bioconductor Rust → Rust-Bio C++ → Bio++ Julia → Bio. Julia Java. Script → Bio. JS ADP
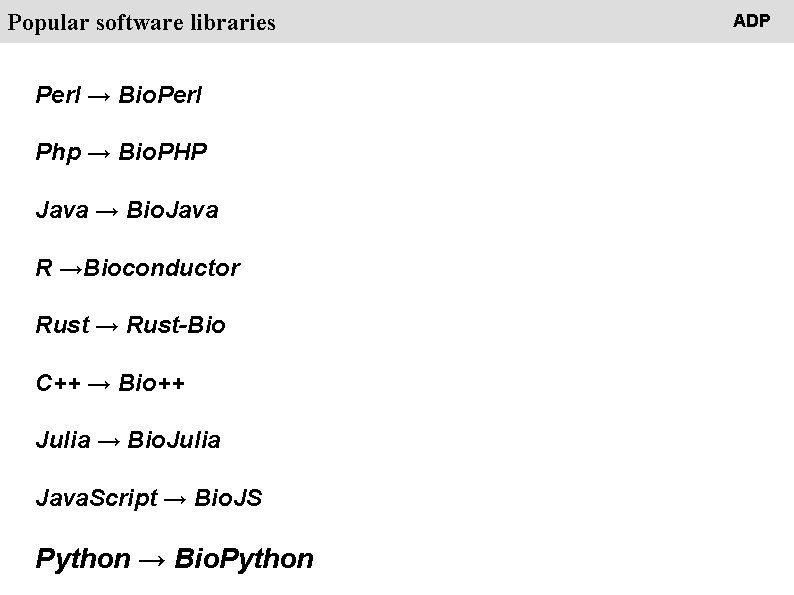
People Popular software libraries Perl → Bio. Perl Php → Bio. PHP Java → Bio. Java R →Bioconductor Rust → Rust-Bio C++ → Bio++ Julia → Bio. Julia Java. Script → Bio. JS Python → Bio. Python ADP
People Popular software libraries Perl → Bio. Perl Php → Bio. PHP Java → Bio. Java R →Bioconductor Rust → Rust-Bio C++ → Bio++ Julia → Bio. Julia Java. Script → Bio. JS Python → Bio. Python, but also Cogent 3, bioconda ADP
People Popular software libraries ADP Perl → Bio. Perl Php → Bio. PHP Java → Bio. Java R →Bioconductor Never restrict yourself only to Bio* libraries Rust → Rust-Bio C++ → Bio++ Julia → Bio. Julia Java. Script → Bio. JS Python → Bio. Python, but also Cogent 3, bioconda
People Popular software libraries ADP Perl → Bio. Perl Php → Bio. PHP Java → Bio. Java R →Bioconductor Never restrict yourself only to Bio* libraries Rust → Rust-Bio C++ → Bio++ Julia → Bio. Julia Java. Script → Bio. JS Python → Bio. Python, but also Cogent 3, bioconda

People Popular software libraries ADP Python → Bio. Python, but also Cogent 3, bioconda Frequently solving bioinformatic problem also means that you will use some custom, small libraries (often with multiple bugs)
People Popular software libraries ADP Python → Bio. Python, but also py. Cogent, bioconda Frequently solving bioinformatic problem also means that you will use some custom, small libraries (often with multiple bugs) For statistics and machine learning: LIBSVM

People Popular software libraries ADP
People Popular software libraries ADP

People Popular software libraries ADP

People Popular software libraries Cogent 3 (as an alternative to biopython) ADP
People Popular software libraries ADP
People Most important bioinformatics databases ADP

People Most important bioinformatics databases https: //www. uniprot. org ADP

People Most important bioinformatics databases ADP
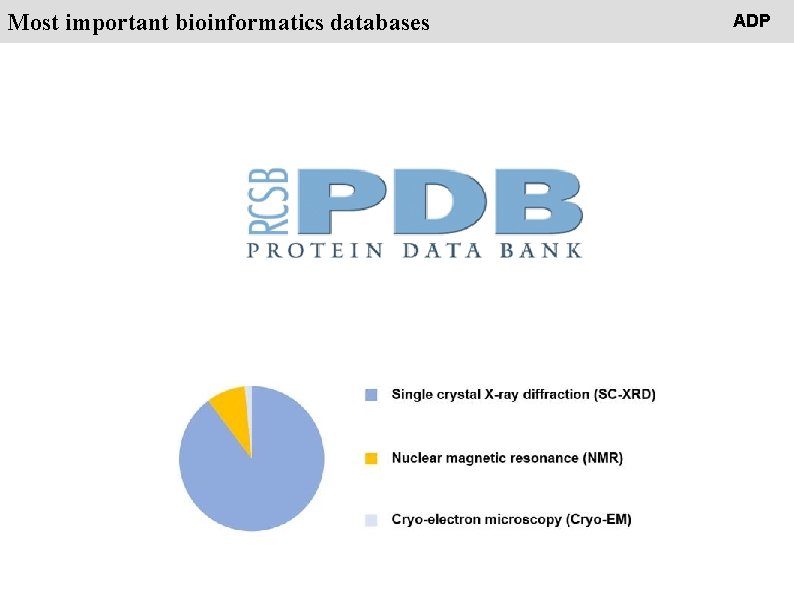
People Most important bioinformatics databases ADP
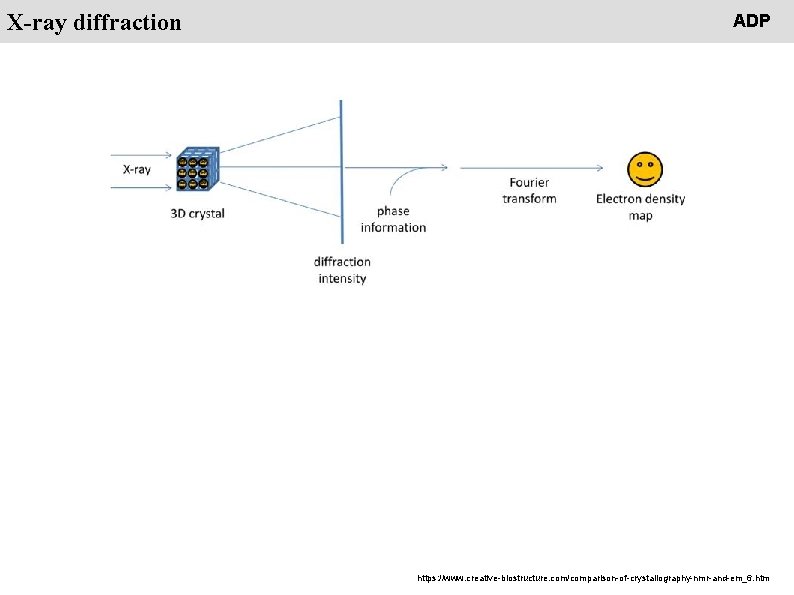
People X-ray diffraction ADP https: //www. creative-biostructure. com/comparison-of-crystallography-nmr-and-em_6. htm
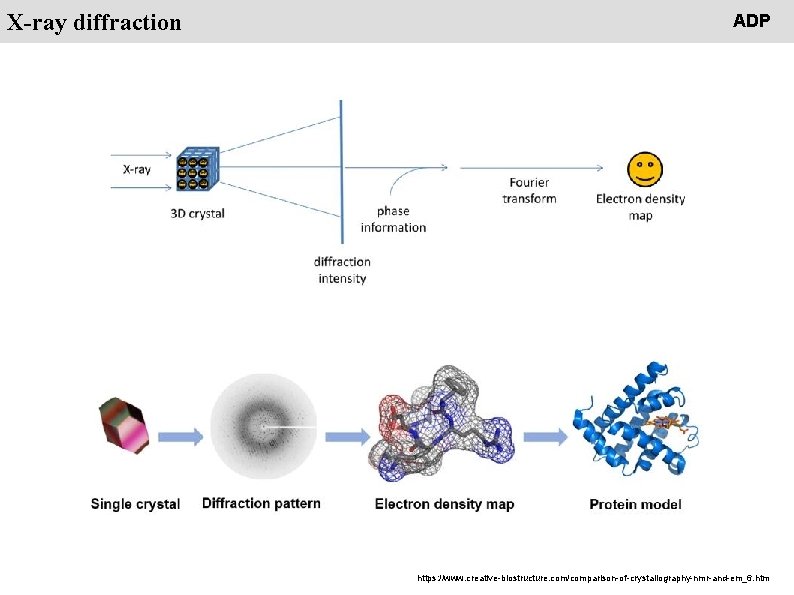
People X-ray diffraction ADP https: //www. creative-biostructure. com/comparison-of-crystallography-nmr-and-em_6. htm
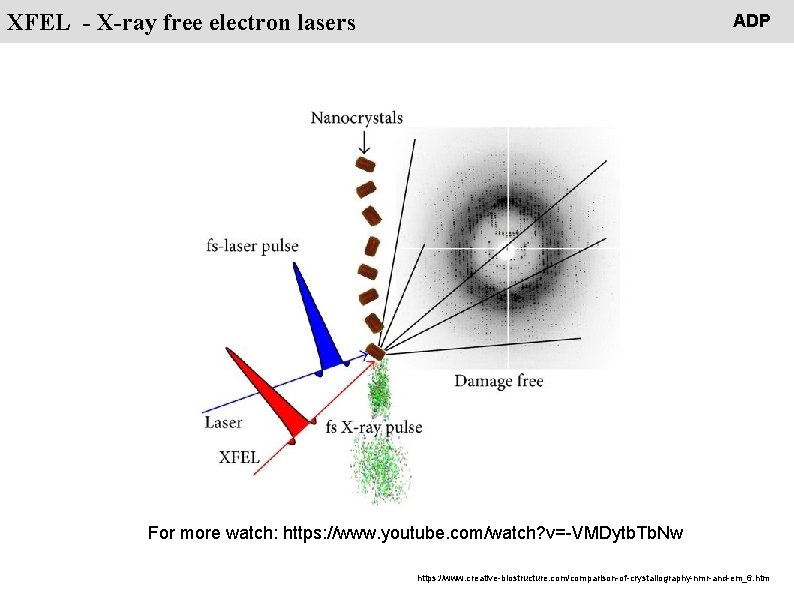
People XFEL - X-ray free electron lasers ADP For more watch: https: //www. youtube. com/watch? v=-VMDytb. Tb. Nw https: //www. creative-biostructure. com/comparison-of-crystallography-nmr-and-em_6. htm
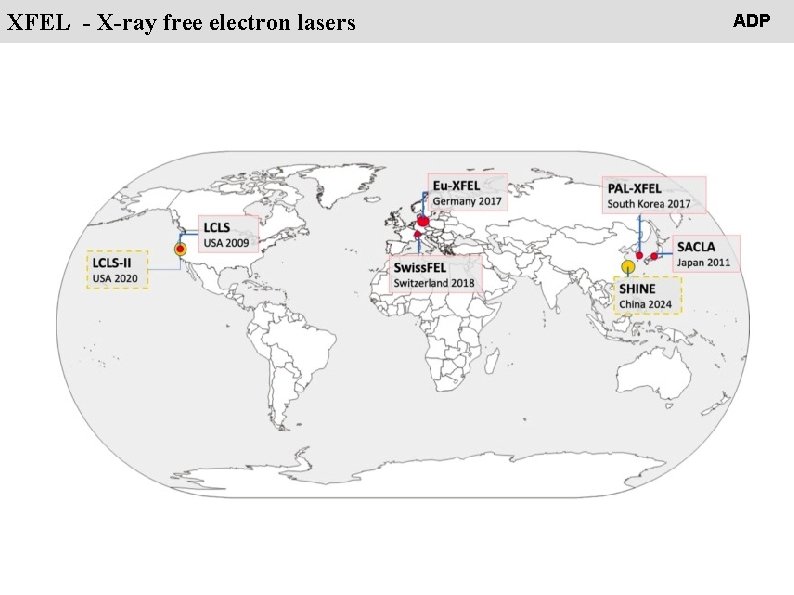
People XFEL - X-ray free electron lasers ADP
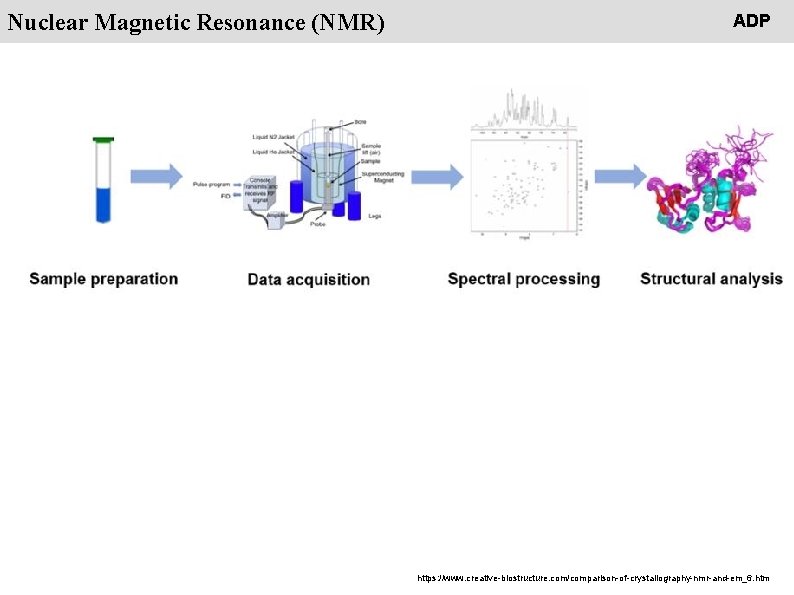
People Nuclear Magnetic Resonance (NMR) ADP https: //www. creative-biostructure. com/comparison-of-crystallography-nmr-and-em_6. htm
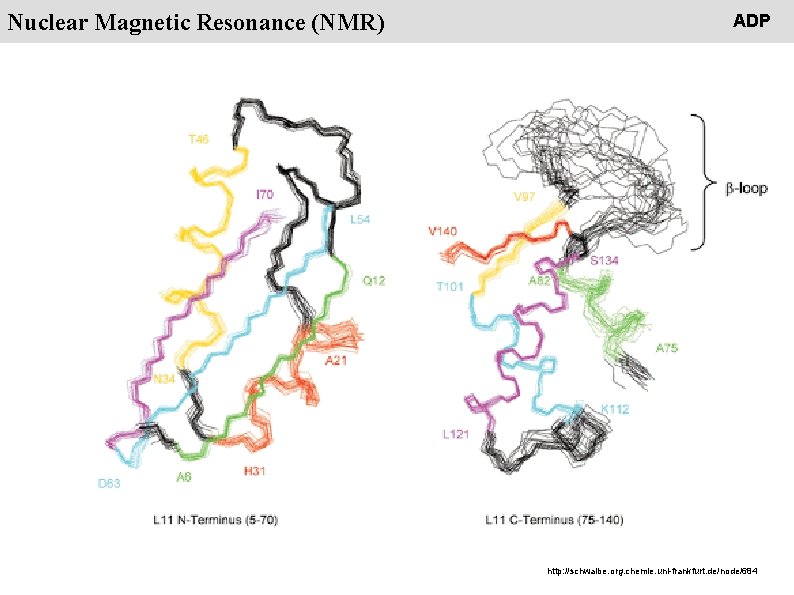
People Nuclear Magnetic Resonance (NMR) ADP http: //schwalbe. org. chemie. uni-frankfurt. de/node/684
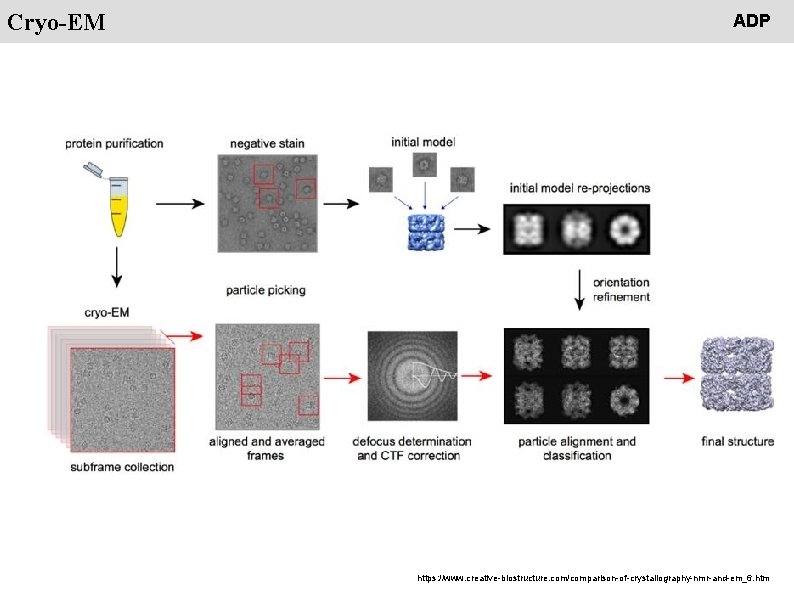
People Cryo-EM ADP https: //www. creative-biostructure. com/comparison-of-crystallography-nmr-and-em_6. htm
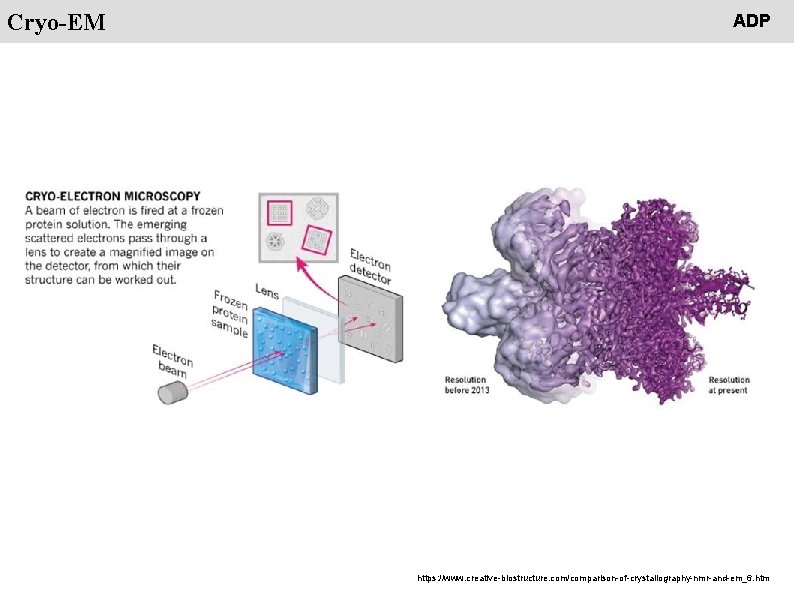
People Cryo-EM ADP https: //www. creative-biostructure. com/comparison-of-crystallography-nmr-and-em_6. htm

People Most important bioinformatics databases https: //www. rcsb. org/ ADP
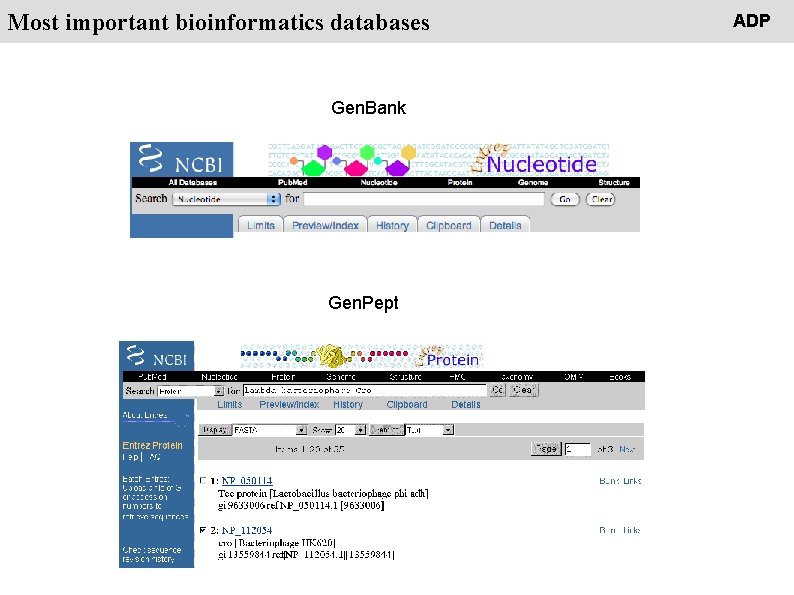
People Most important bioinformatics databases Gen. Bank Gen. Pept ADP
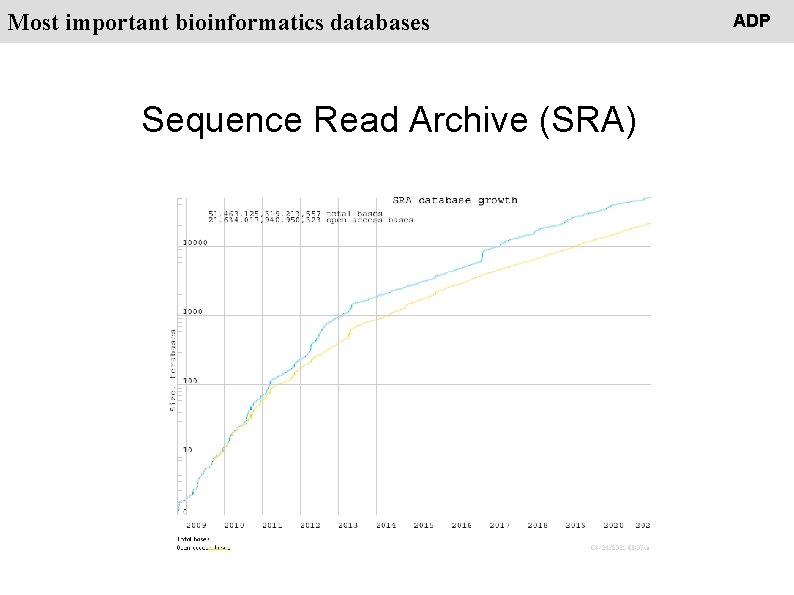
People Most important bioinformatics databases Sequence Read Archive (SRA) ADP

People Most important bioinformatics databases Sequence Read Archive (SRA) stores raw sequence data from "next-generation" sequencing technologies including Illumina, 454, Ion. Torrent, Complete Genomics, Pac. Bio and Oxford. Nanopores ADP

People Most important bioinformatics databases Sequence Read Archive (SRA) stores raw sequence data from "next-generation" sequencing technologies including Illumina, 454, Ion. Torrent, Complete Genomics, Pac. Bio and Oxford. Nanopores SRA = NGS data ADP
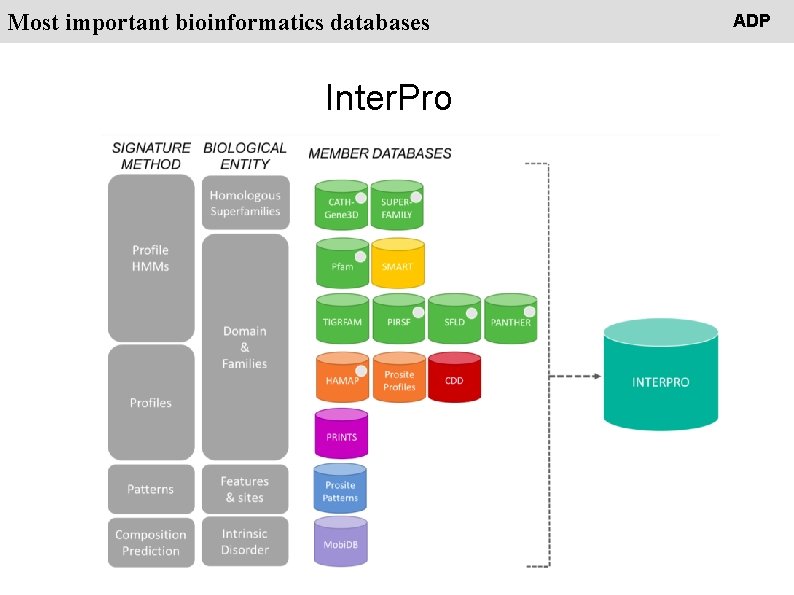
People Most important bioinformatics databases Inter. Pro ADP
People Most important bioinformatics databases ADP
People Most important bioinformatics databases ADP
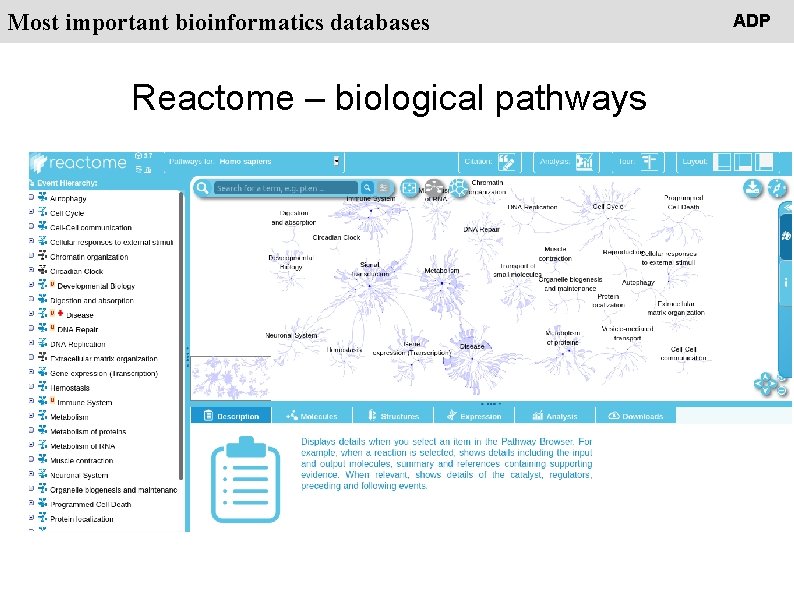
People Most important bioinformatics databases Reactome – biological pathways ADP

Thank you for your time and See you at the next lecture Any other questions & comments lukaskoz@mimuw. edu. pl
- Slides: 48