Architecture Exploration of FPGA based Accelerators for Bioinformatics
Architecture Exploration of FPGA based Accelerators for Bioinformatics Thesis Presentation B. Sharat Chandra Varma Amarnath Shashi Khosla School of Information Technology, varma@cse. iitd. ac. in Supervisors Dr. Kolin Paul Department of Computer Science and Engineering kolin@cse. iitd. ac. in Prof. M. Balakrishnan Department of Computer Science and Engineering mbala@cse. iitd. ac. in
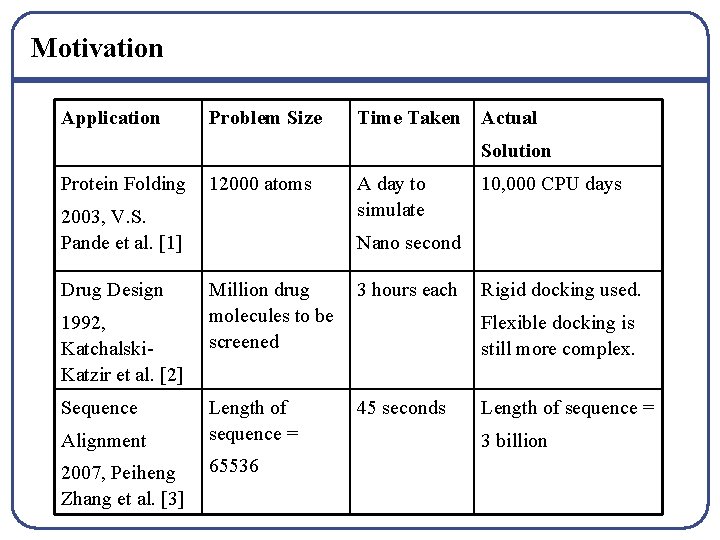
Motivation Application Problem Size Time Taken Actual Solution Protein Folding 12000 atoms 2003, V. S. Pande et al. [1] Drug Design 1992, Katchalski. Katzir et al. [2] Sequence Alignment 2007, Peiheng Zhang et al. [3] A day to simulate 10, 000 CPU days Nano second Million drug molecules to be screened 3 hours each Length of sequence = 45 seconds 65536 Rigid docking used. Flexible docking is still more complex. Length of sequence = 3 billion
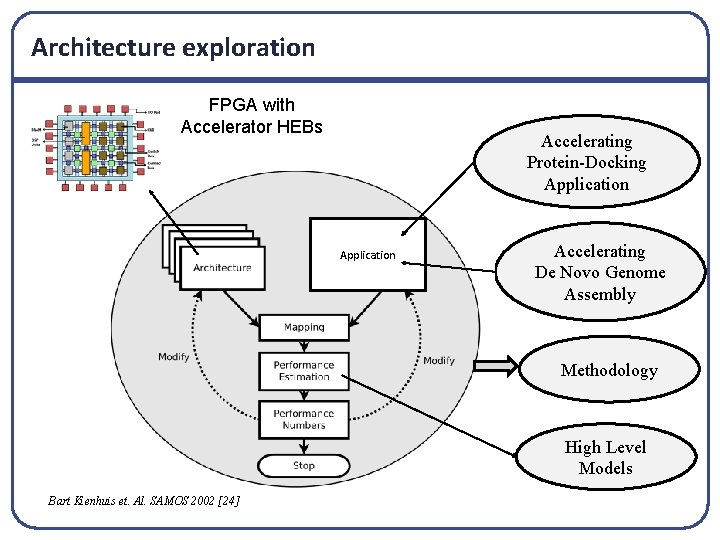
Architecture exploration FPGA with Accelerator HEBs Accelerating Protein-Docking Application Accelerating De Novo Genome Assembly Methodology High Level Models Bart Kienhuis et. Al. SAMOS 2002 [24]
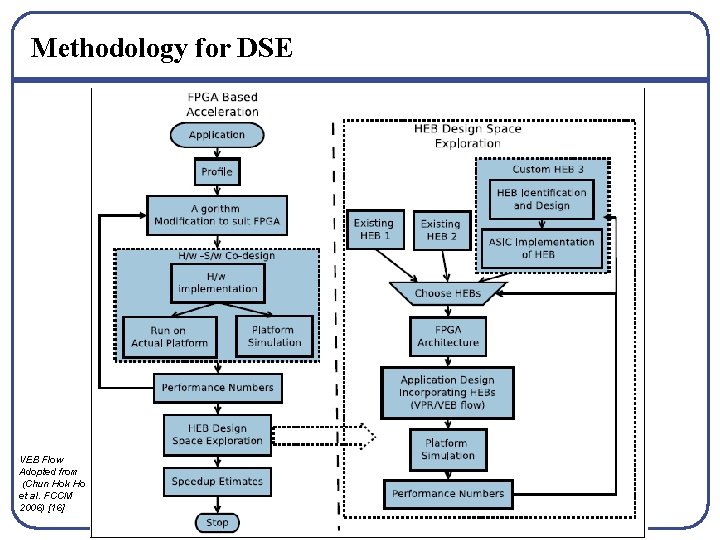
Methodology for DSE VEB Flow Adopted from (Chun Hok Ho et al. FCCM 2006) [16]
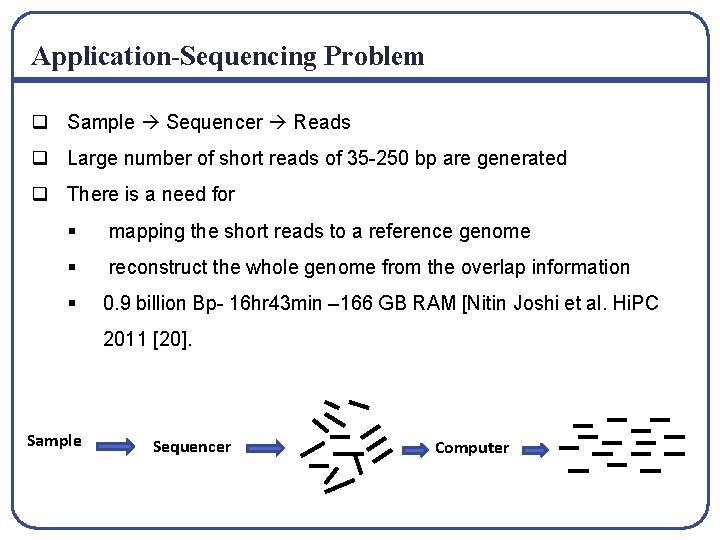
Application-Sequencing Problem q Sample Sequencer Reads q Large number of short reads of 35 -250 bp are generated q There is a need for § mapping the short reads to a reference genome § reconstruct the whole genome from the overlap information § 0. 9 billion Bp- 16 hr 43 min – 166 GB RAM [Nitin Joshi et al. Hi. PC 2011 [20]. Sample Sequencer Computer
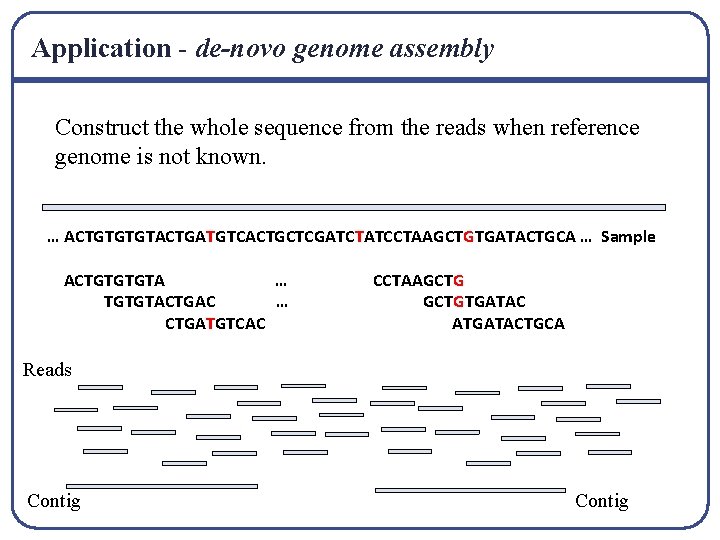
Application - de-novo genome assembly Construct the whole sequence from the reads when reference genome is not known. … ACTGTGTGTACTGATGTCACTGCTCGATCTATCCTAAGCTGTGATACTGCA … Sample ACTGTGTGTA … TGTGTACTGAC … CTGATGTCAC CCTAAGCTGTGATAC ATGATACTGCA Reads Contig
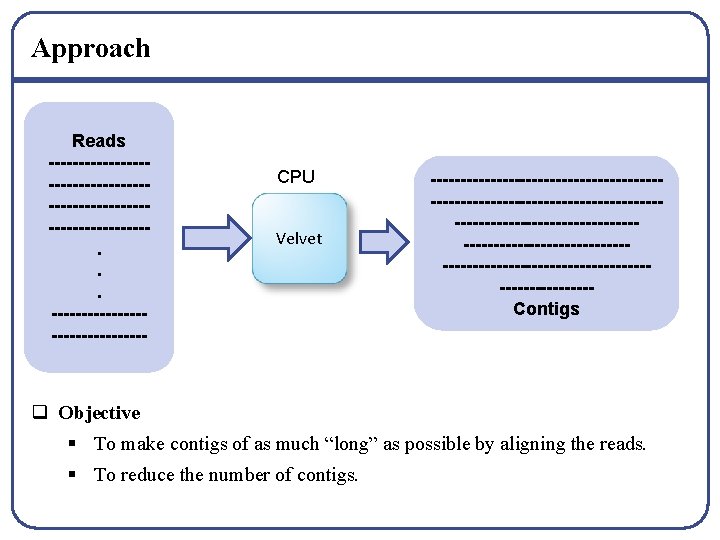
Approach Reads --------------------------------. . . ---------------- CPU Velvet -------------------------------------------------------------------------------------------Contigs q Objective § To make contigs of as much “long” as possible by aligning the reads. § To reduce the number of contigs.
![Different Models Parameters q C-Model using Mapsembler [17] § Overall Speed-up § Quality of Different Models Parameters q C-Model using Mapsembler [17] § Overall Speed-up § Quality of](http://slidetodoc.com/presentation_image_h/05bc7734dec205df7c90fcd6c38dc361/image-9.jpg)
Different Models Parameters q C-Model using Mapsembler [17] § Overall Speed-up § Quality of Output § Simulation of Large Genomes § Initial Algorithm Changes q System C-Model § Threshold Variation § Pre-Filter Design § Simulation with HEBs § Effect of pre-filter on speedup q VHDL Model § Clock Speed § Number of PEs in FPGA § HEB design
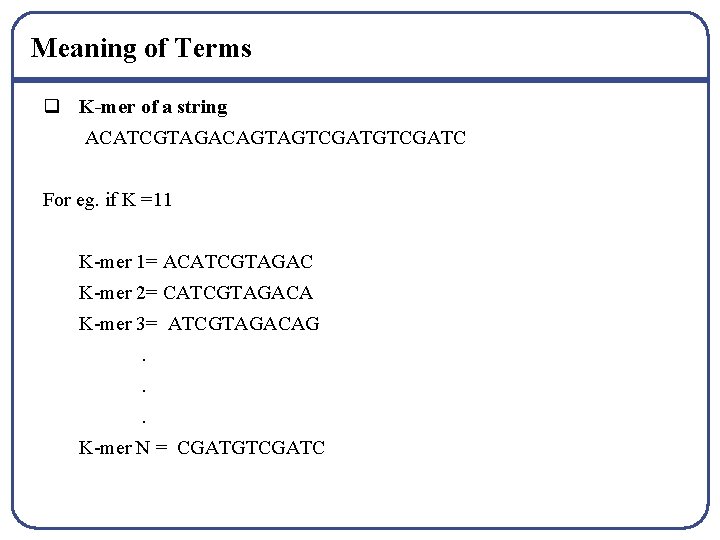
Meaning of Terms q K-mer of a string ACATCGTAGACAGTAGTCGATC For eg. if K =11 K-mer 1= ACATCGTAGAC K-mer 2= CATCGTAGACA K-mer 3= ATCGTAGACAG. . . K-mer N = CGATGTCGATC

Meaning of Extension q Starter ACATCGTAGACAGTAGTCGATC q Read TGGATGATAGCATCGTAGACACA Extend starter ACATCGTAGACAGTAGTCGATGTCGATC ACATCGTAGACAGTAGTCGATC TGGATGATAACATCGTAGACAGT TGGATGATAACATCGTAGACAGT Cycle 9 1 2 3 q Extended Starter TGGATGATAACATCGTAGACAGTAGTCGATC q Extension at the edges. Either right or left.
![MAPSEMBLER[25] for Assembly – C Model ---------------------------------------------. Reads. ------------------------------------------------------------- Random reads as Starters ----------------------- MAPSEMBLER[25] for Assembly – C Model ---------------------------------------------. Reads. ------------------------------------------------------------- Random reads as Starters -----------------------](http://slidetodoc.com/presentation_image_h/05bc7734dec205df7c90fcd6c38dc361/image-12.jpg)
MAPSEMBLER[25] for Assembly – C Model ---------------------------------------------. Reads. ------------------------------------------------------------- Random reads as Starters ----------------------- Kmer 1: (frag 10, pos 3) Kmer 2: frag 2, pos 3)(frag 8, pos 1). -----------------------------------Extended Starter. . . -------- Delete Read Hash Table Update Hash. . Kmer n-1: (frag 9, pos 3) Kmer n: frag 3, pos 3)(frag 8, pos 1)
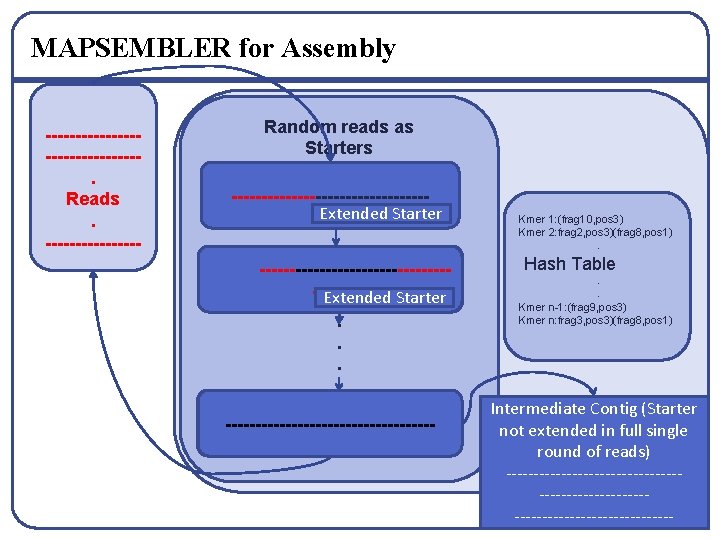
MAPSEMBLER for Assembly ---------------. Reads. -------- Random reads as Starters ----------------Extended Starter -----------------------------------------Extended Starter. . . ------------------------- Kmer 1: (frag 10, pos 3) Kmer 2: frag 2, pos 3)(frag 8, pos 1). Hash Table. . Kmer n-1: (frag 9, pos 3) Kmer n: frag 3, pos 3)(frag 8, pos 1) Intermediate Contig (Starter not extended in full single round of reads) ----------------------------------------
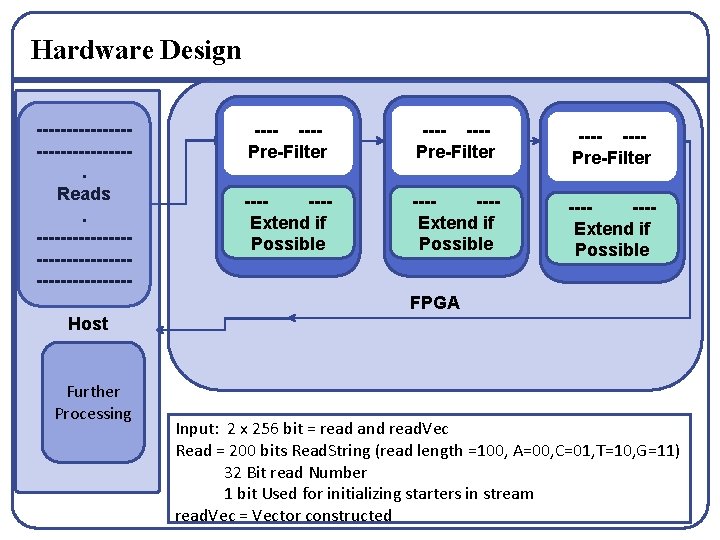
Hardware Design ---------------. Reads. ----------------------- ---Pre-Filter ------Extend if Possible ------Extend if Possible FPGA Host Further Processing Input: 2 x 256 bit = read and read. Vec Read = 200 bits Read. String (read length =100, A=00, C=01, T=10, G=11) 32 Bit read Number 1 bit Used for initializing starters in stream read. Vec = Vector constructed
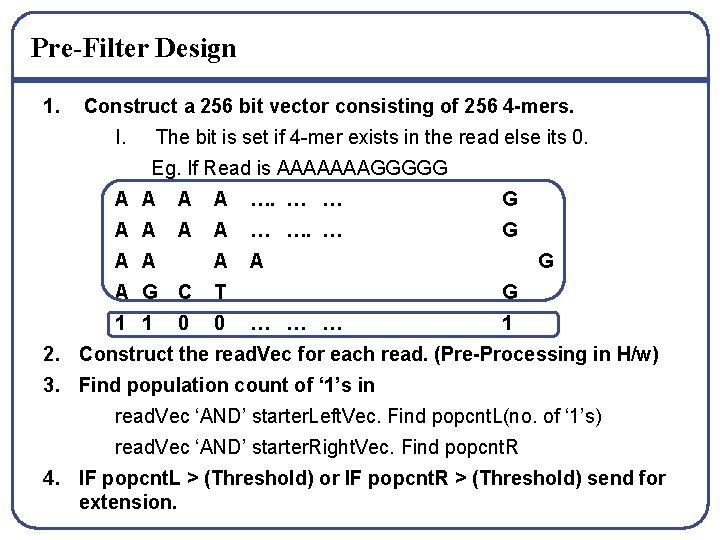
Pre-Filter Design 1. Construct a 256 bit vector consisting of 256 4 -mers. I. The bit is set if 4 -mer exists in the read else its 0. Eg. If Read is AAAAAAAGGGGG A A …. … … G A A … …. … G A A A G C T 1 1 0 0 G G … … … 1 2. Construct the read. Vec for each read. (Pre-Processing in H/w) 3. Find population count of ‘ 1’s in read. Vec ‘AND’ starter. Left. Vec. Find popcnt. L(no. of ‘ 1’s) read. Vec ‘AND’ starter. Right. Vec. Find popcnt. R 4. IF popcnt. L > (Threshold) or IF popcnt. R > (Threshold) send for extension.

Hardware Design q Read is represented in binary coded format § A=“ 00”, C= “ 01”, T=“ 10” and G=“ 11” q We do not store the whole starter. § Store the left end and right end of starter equivalent to read length. § Eg. ACTGCTGTGTGTGTGATGTACTGCA if Read length = 9 § starter. Left § starter. Right = TGTACTGCA § For first tiime we store the read as both starter. Left and starter. Right = ACTGCTGTG q We consider precheck. § It checks if there is possibility of extension. § Shift and extend if possible.
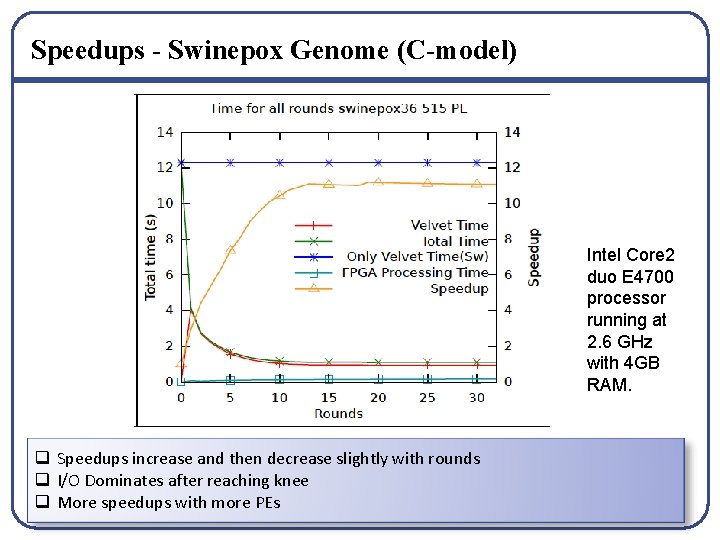
Speedups - Swinepox Genome (C-model) Intel Core 2 duo E 4700 processor running at 2. 6 GHz with 4 GB RAM. q Speedups increase and then decrease slightly with rounds q I/O Dominates after reaching knee q More speedups with more PEs
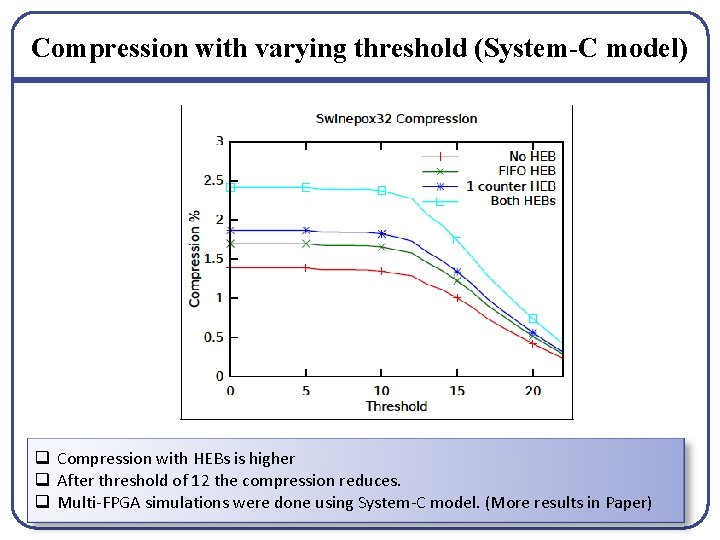
Compression with varying threshold (System-C model) q Compression with HEBs is higher q After threshold of 12 the compression reduces. q Multi-FPGA simulations were done using System-C model. (More results in Paper)
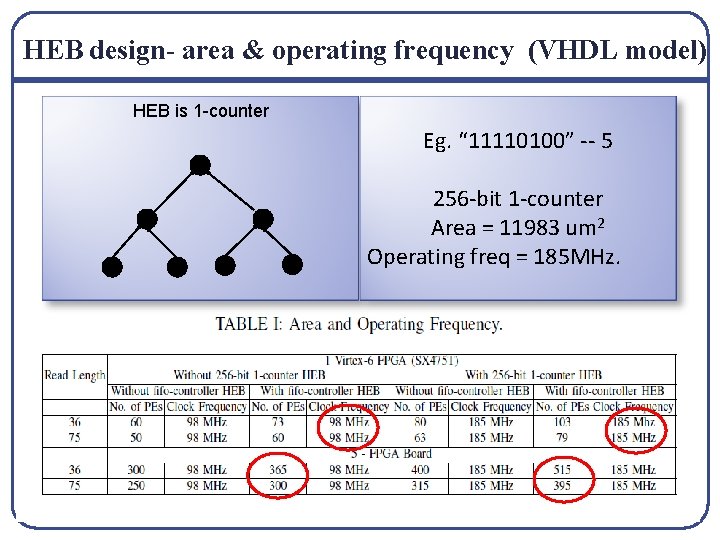
HEB design- area & operating frequency (VHDL model) HEB is 1 -counter Eg. “ 11110100” -- 5 256 -bit 1 -counter Area = 11983 um 2 Operating freq = 185 MHz.
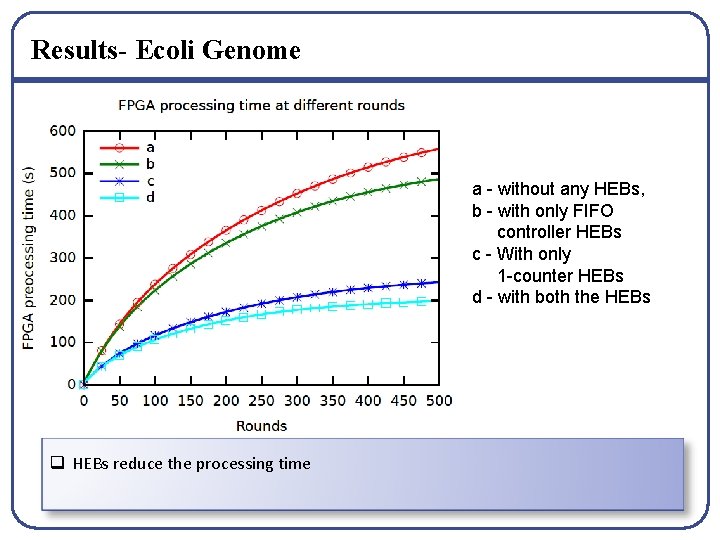
Results- Ecoli Genome a - without any HEBs, b - with only FIFO controller HEBs c - With only 1 -counter HEBs d - with both the HEBs q HEBs reduce the processing time
- Slides: 20