Approaches to describe the shade avoidance response genetic

Approaches to describe the shade avoidance response genetic network 05 -16 -07
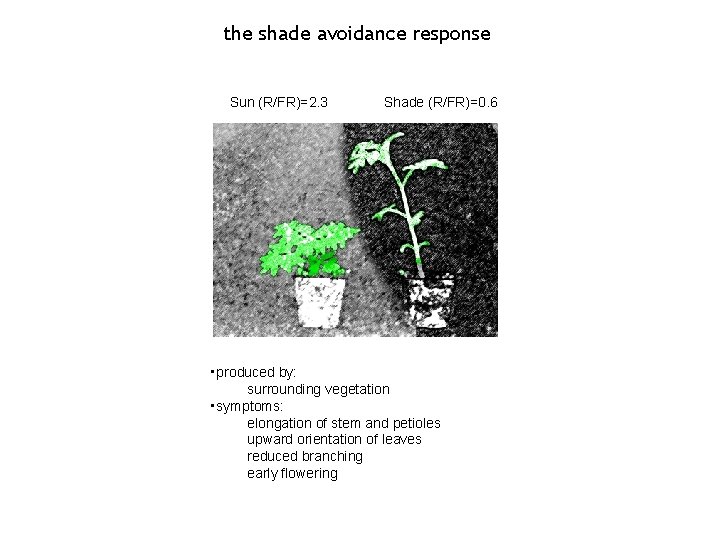
the shade avoidance response Sun (R/FR)=2. 3 Shade (R/FR)=0. 6 • produced by: surrounding vegetation • symptoms: elongation of stem and petioles upward orientation of leaves reduced branching early flowering
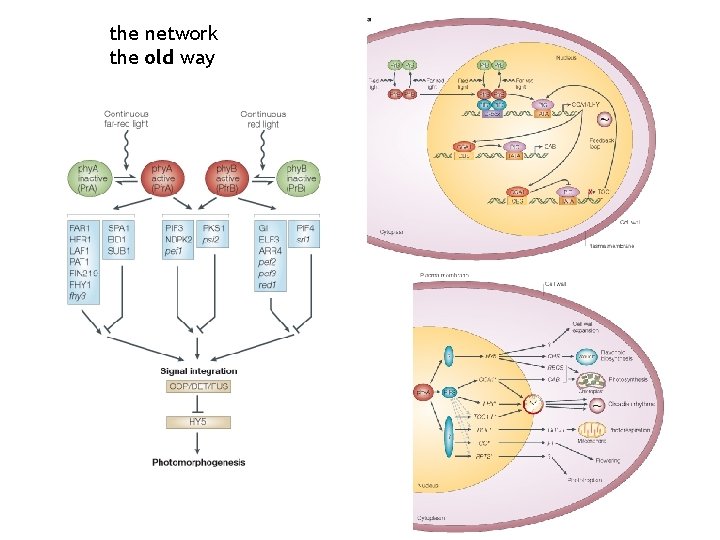
the network the old way
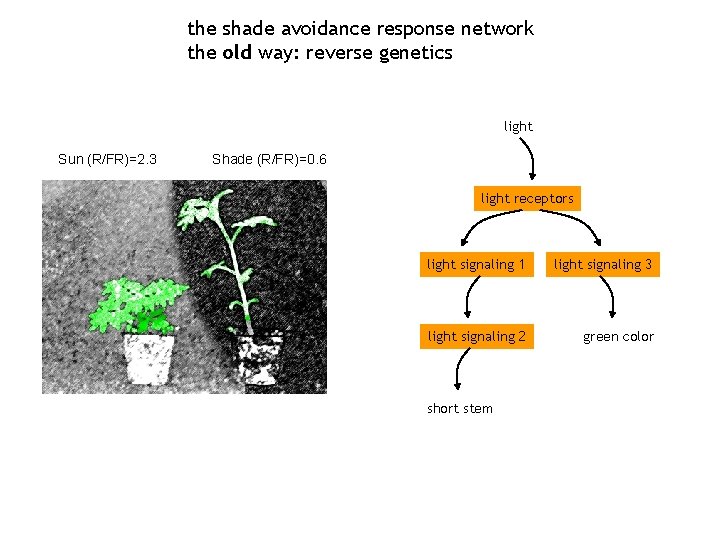
the shade avoidance response network the old way: reverse genetics light Sun (R/FR)=2. 3 Shade (R/FR)=0. 6 light receptors light signaling 1 light signaling 3 light signaling 2 green color short stem
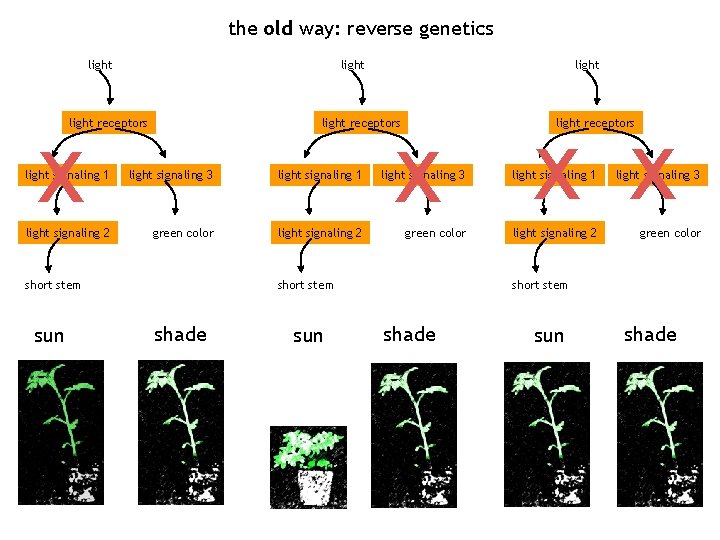
the old way: reverse genetics light x light receptors light x x x light receptors light signaling 1 light signaling 3 light signaling 2 green color short stem sun short stem shade sun shade
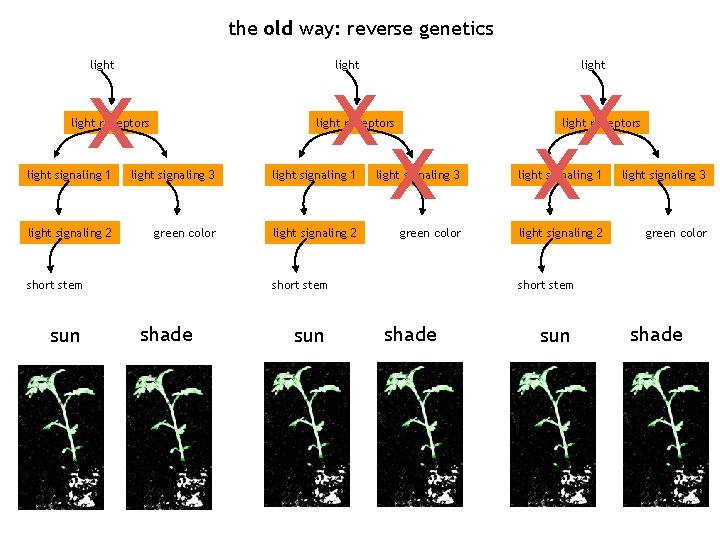
the old way: reverse genetics x light receptors x x light receptors light signaling 1 light signaling 3 light signaling 2 green color short stem sun shade sun short stem shade sun shade
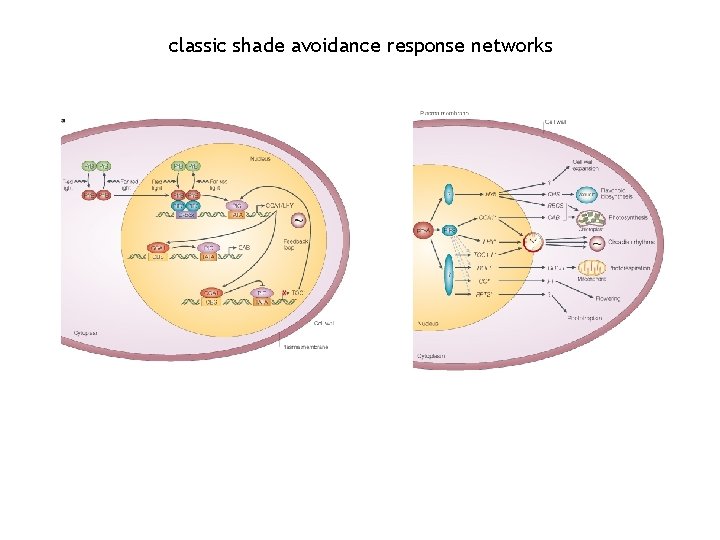
classic shade avoidance response networks
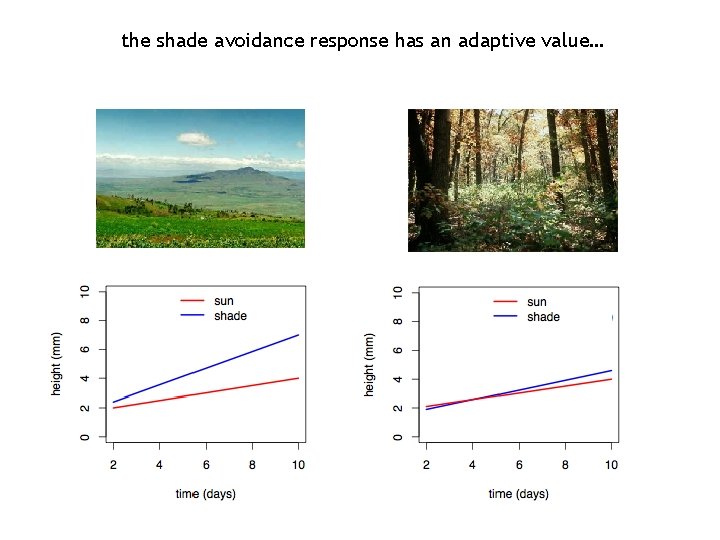
the shade avoidance response has an adaptive value…
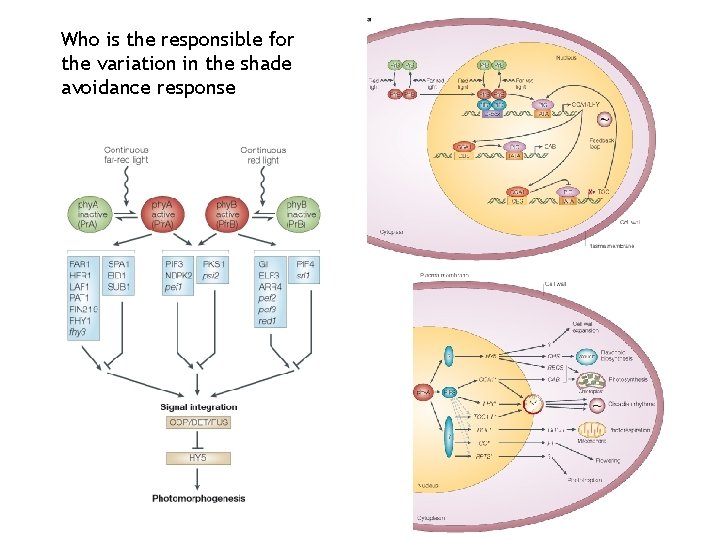
Who is the responsible for the variation in the shade avoidance response
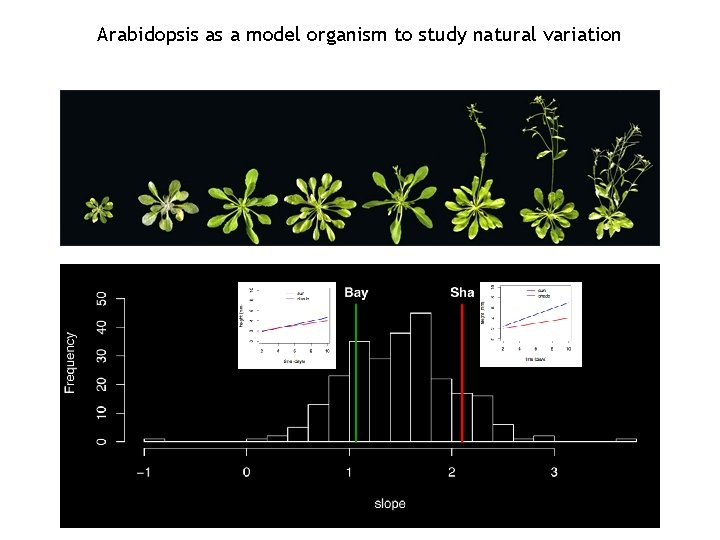
Arabidopsis as a model organism to study natural variation
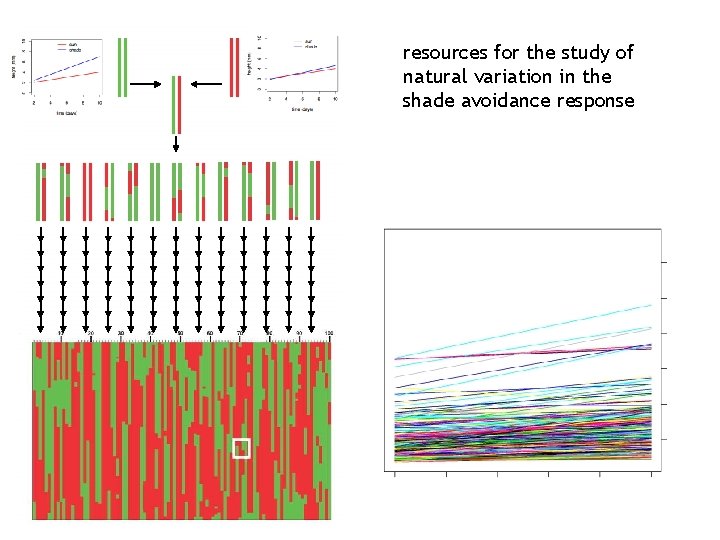
resources for the study of natural variation in the shade avoidance response
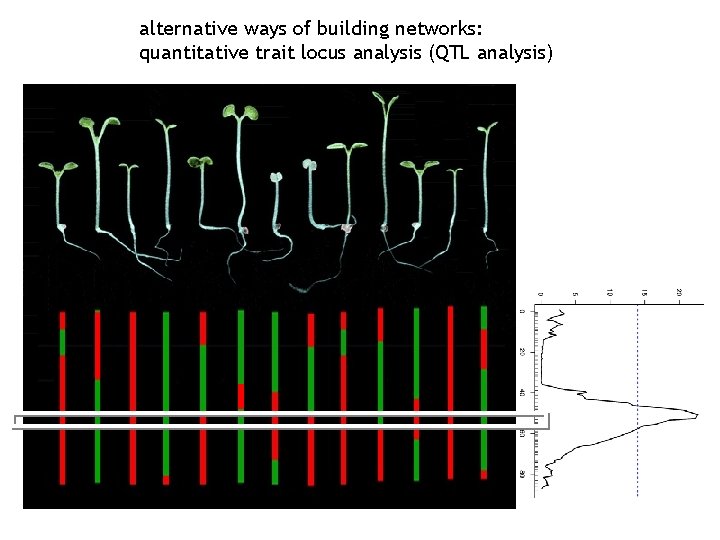
alternative ways of building networks: quantitative trait locus analysis (QTL analysis)
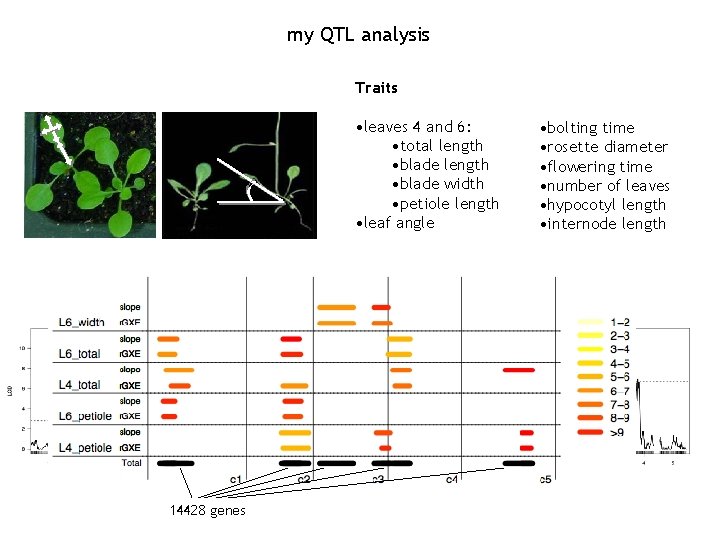
my QTL analysis Traits • leaves 4 and 6: • total length • blade width • petiole length • leaf angle 14428 genes • bolting time • rosette diameter • flowering time • number of leaves • hypocotyl length • internode length
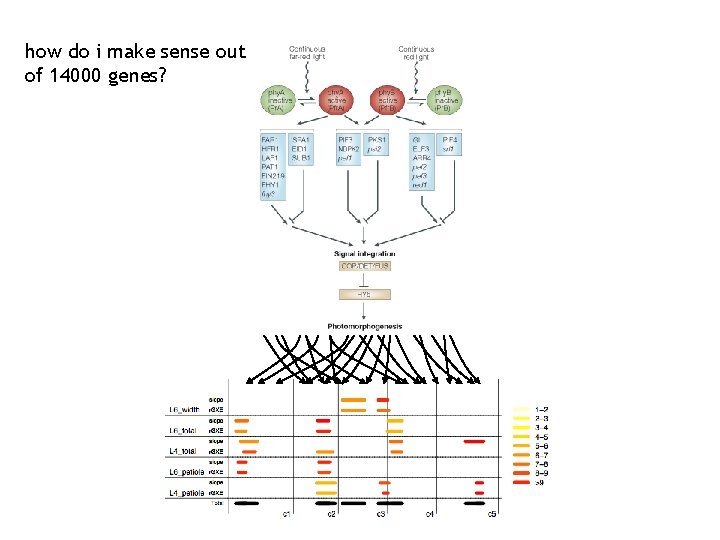
how do i make sense out of 14000 genes?
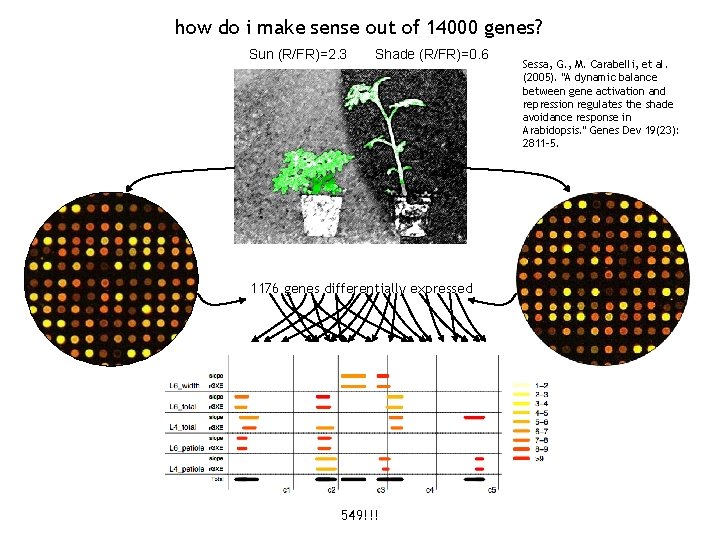
how do i make sense out of 14000 genes? Sun (R/FR)=2. 3 Shade (R/FR)=0. 6 1176 genes differentially expressed 549!!! Sessa, G. , M. Carabelli, et al. (2005). "A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. " Genes Dev 19(23): 2811 -5.
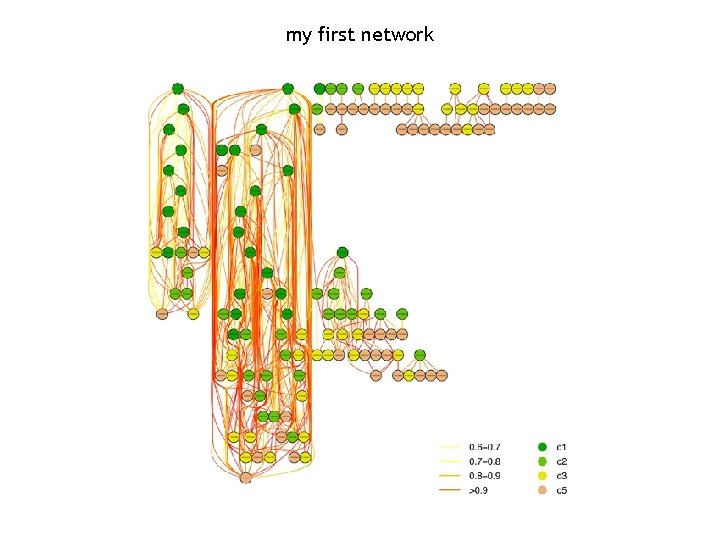
my first network
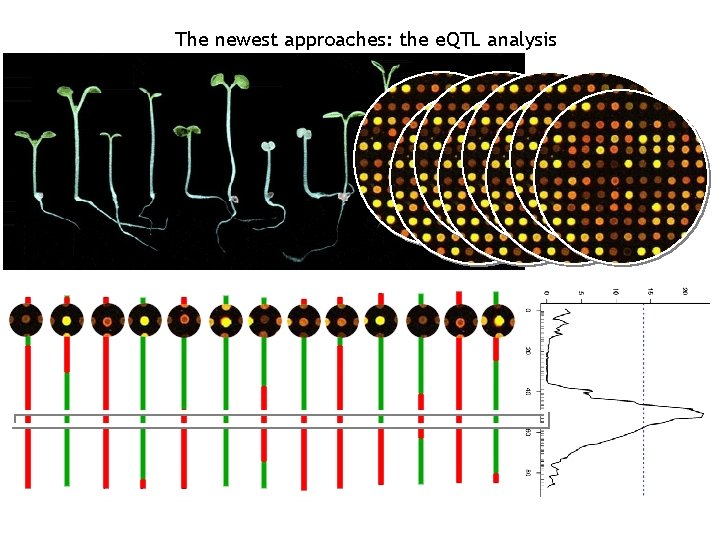
The newest approaches: the e. QTL analysis • Keurentjes, J. J. B. , J. Fu, et al. (2007). "Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. " PNAS 104(5): 1708 -1713. • West, M. A. L. , K. Kim, et al. (2007). "Global e. QTL Mapping Reveals the Complex Genetic Architecture of Transcript. Level Variation in Arabidopsis. " Genetics 175(3): 1441 -1450.
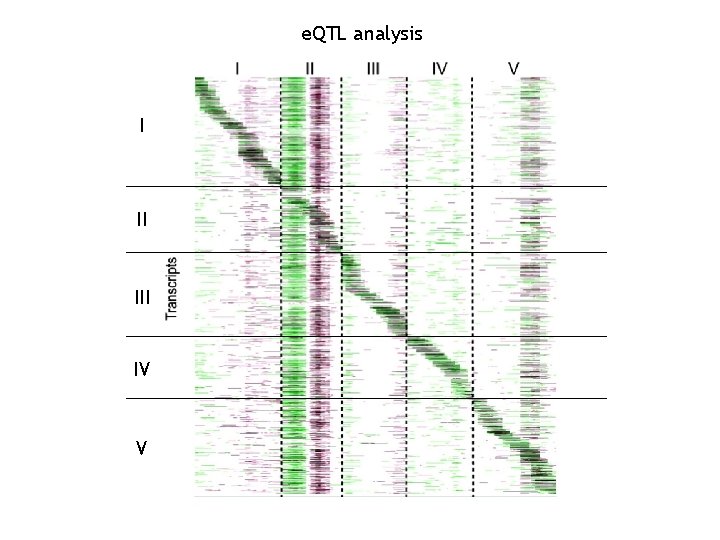
e. QTL analysis I II IV V
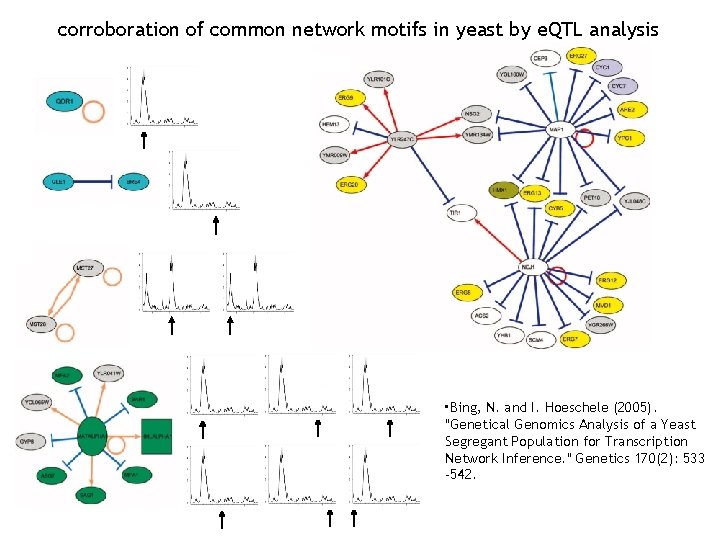
corroboration of common network motifs in yeast by e. QTL analysis • Bing, N. and I. Hoeschele (2005). "Genetical Genomics Analysis of a Yeast Segregant Population for Transcription Network Inference. " Genetics 170(2): 533 -542.
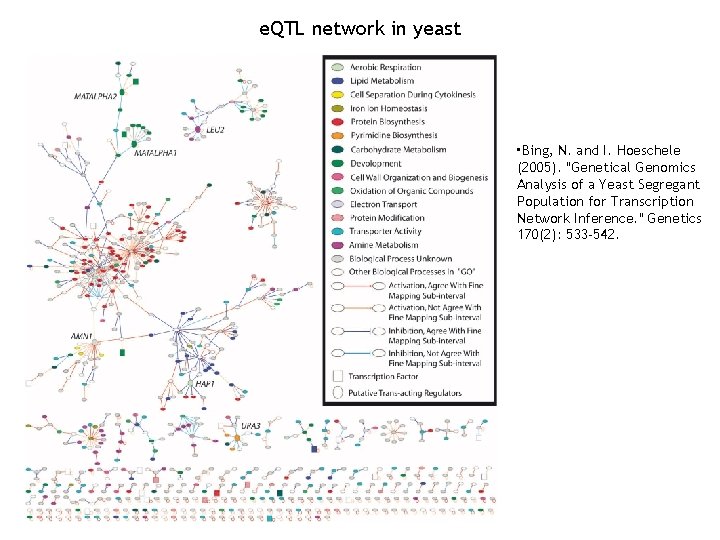
e. QTL network in yeast • Bing, N. and I. Hoeschele (2005). "Genetical Genomics Analysis of a Yeast Segregant Population for Transcription Network Inference. " Genetics 170(2): 533 -542.
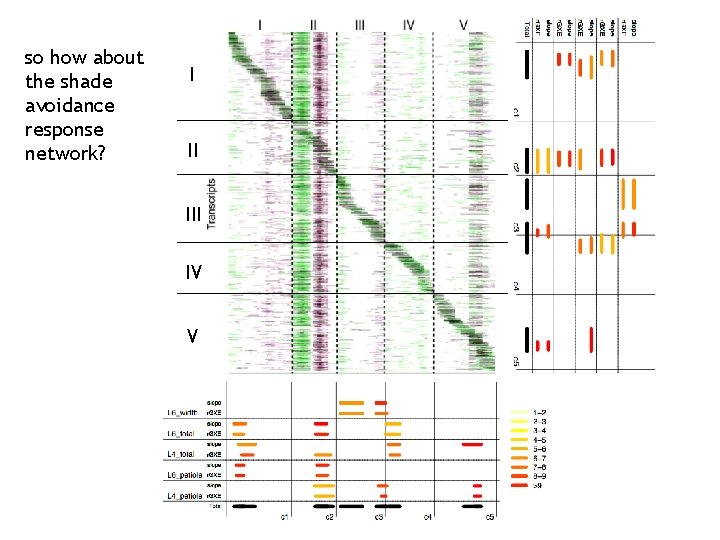
so how about the shade avoidance response network? I II IV V
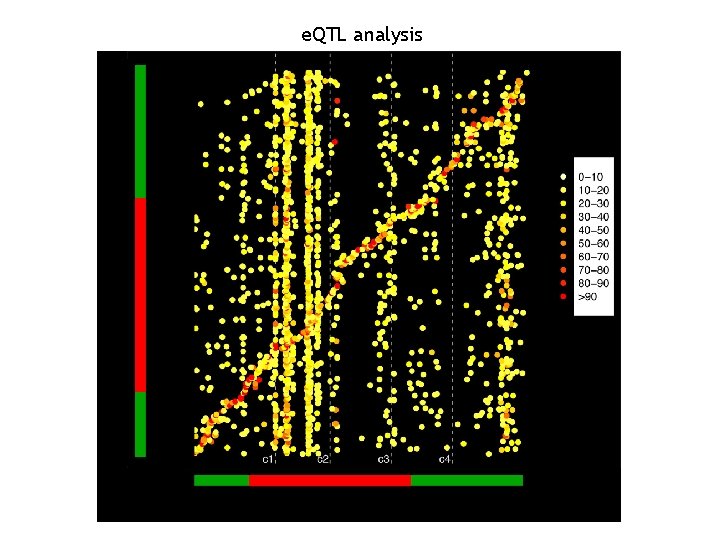
e. QTL analysis
- Slides: 22