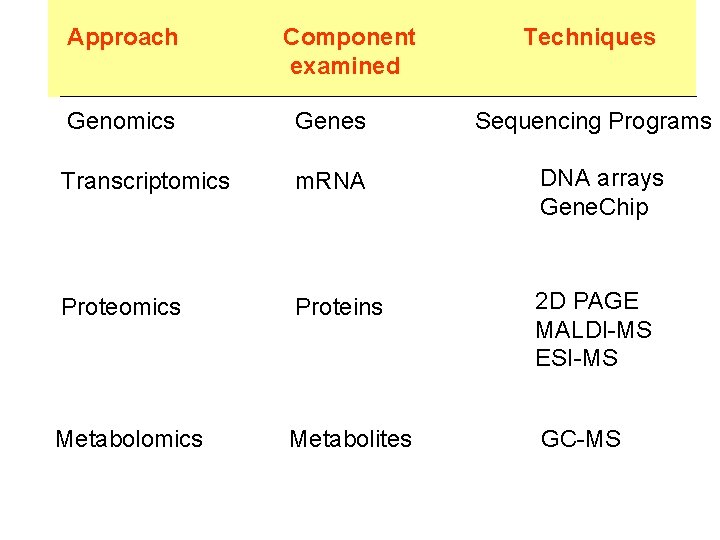
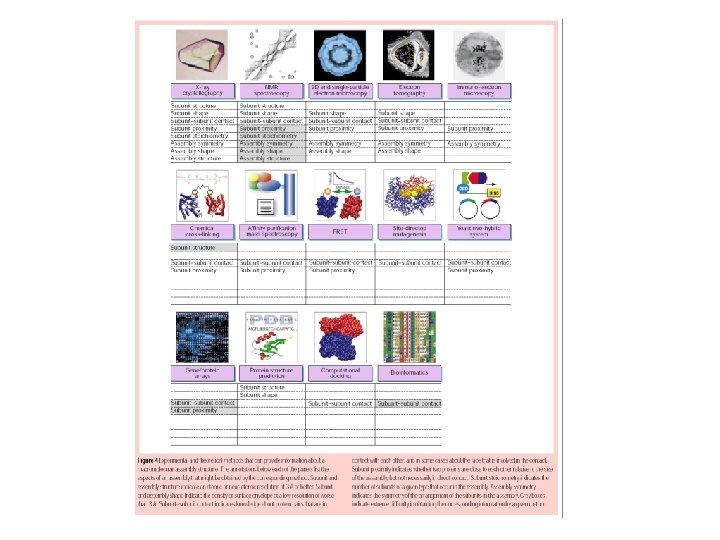
Approach Component examined Techniques Genomics Genes Transcriptomics m

































- Slides: 67

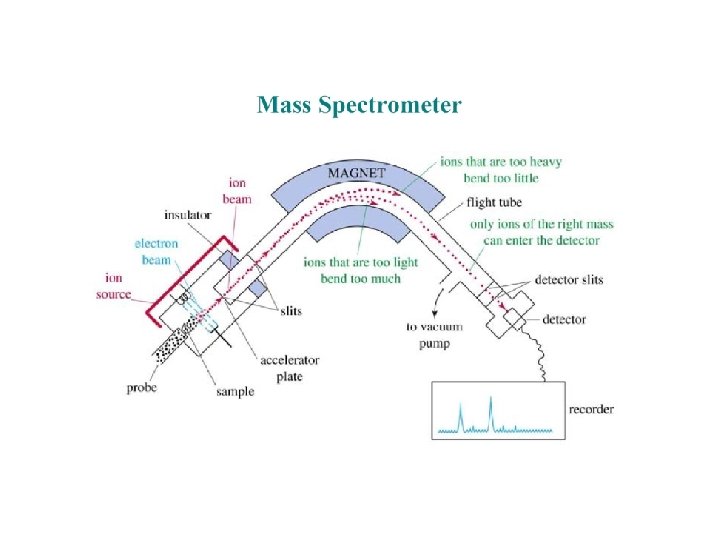
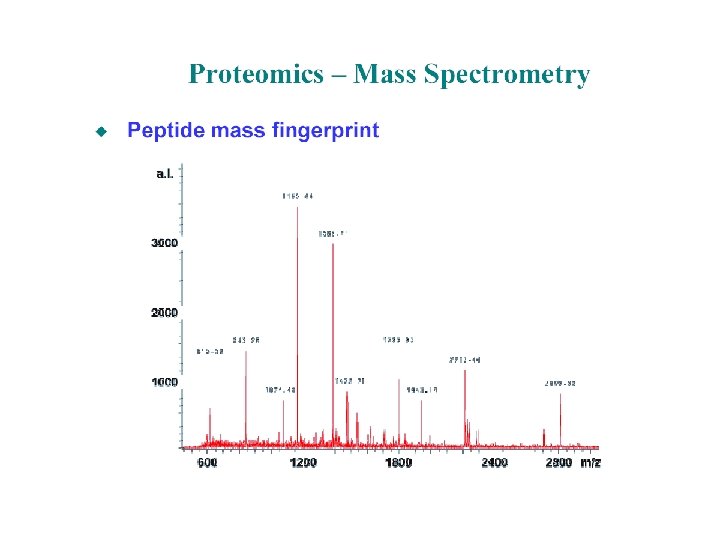
Approach Component examined Techniques Genomics Genes Transcriptomics m. RNA DNA arrays Gene. Chip Proteomics Proteins 2 D PAGE MALDI-MS ESI-MS Metabolomics Metabolites GC-MS Sequencing Programs

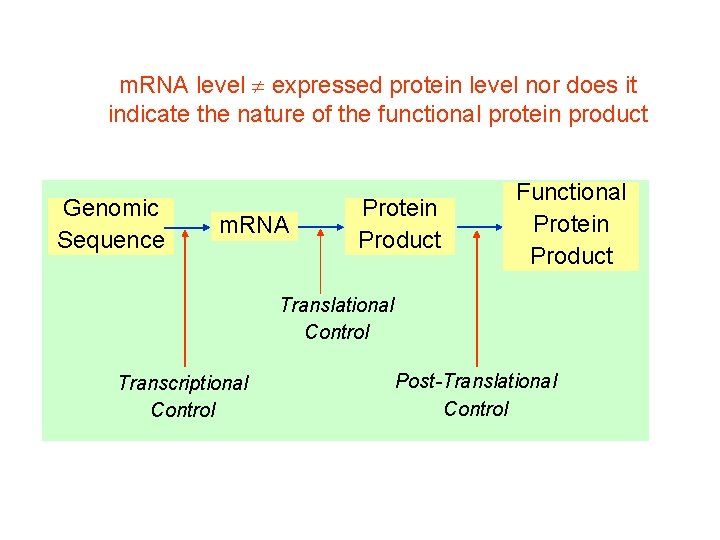
m. RNA level expressed protein level nor does it indicate the nature of the functional protein product Genomic Sequence m. RNA Protein Product Functional Protein Product Translational Control Transcriptional Control Post-Translational Control

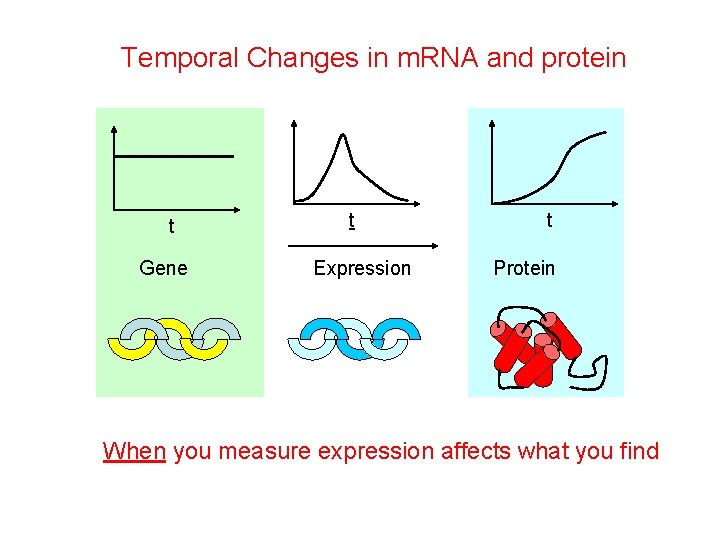
Temporal Changes in m. RNA and protein t Gene t Expression t Protein When you measure expression affects what you find

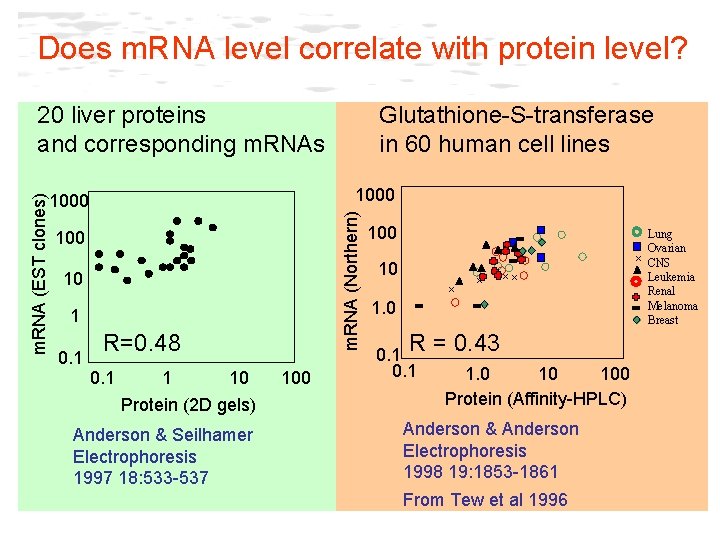
Does m. RNA level correlate with protein level? 1000 100 10 1 0. 1 Glutathione-S-transferase in 60 human cell lines m. RNA (Northern) m. RNA (EST clones) 20 liver proteins and corresponding m. RNAs R=0. 48 0. 1 1 10 Protein (2 D gels) Anderson & Seilhamer Electrophoresis 1997 18: 533 -537 100 x 10 x x xx 1. 0 R = 0. 43 0. 1 1. 0 10 100 Protein (Affinity-HPLC) Anderson & Anderson Electrophoresis 1998 19: 1853 -1861 From Tew et al 1996 Lung Ovarian x CNS Leukemia Renal Melanoma Breast

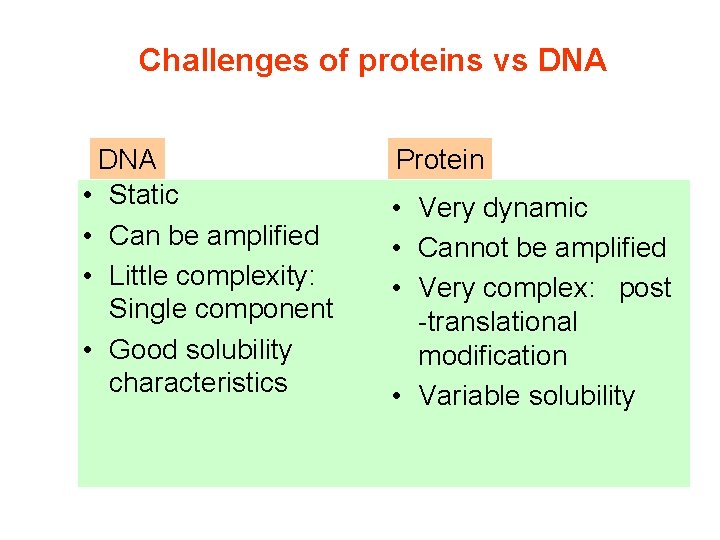
Challenges of proteins vs DNA • Static • Can be amplified • Little complexity: Single component • Good solubility characteristics Protein • Very dynamic • Cannot be amplified • Very complex: post -translational modification • Variable solubility














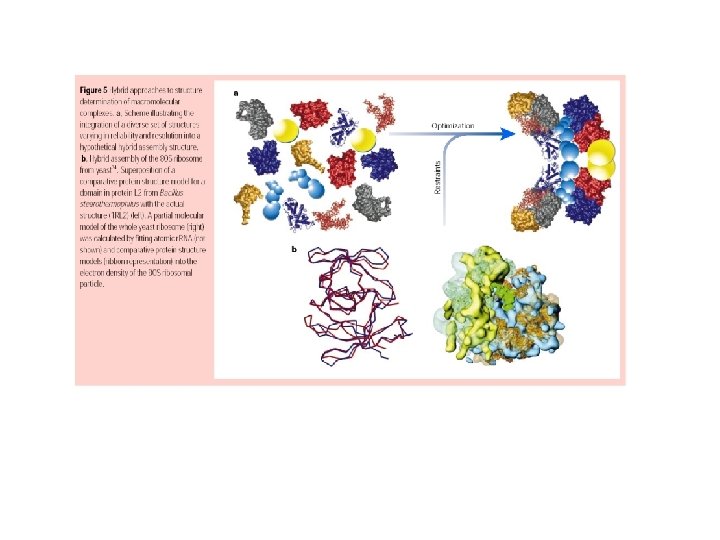
Identifying new protein complexes: Isolation of proteins using: Classical Purification +1 D PAGE Tag Purification +1 D PAGE



















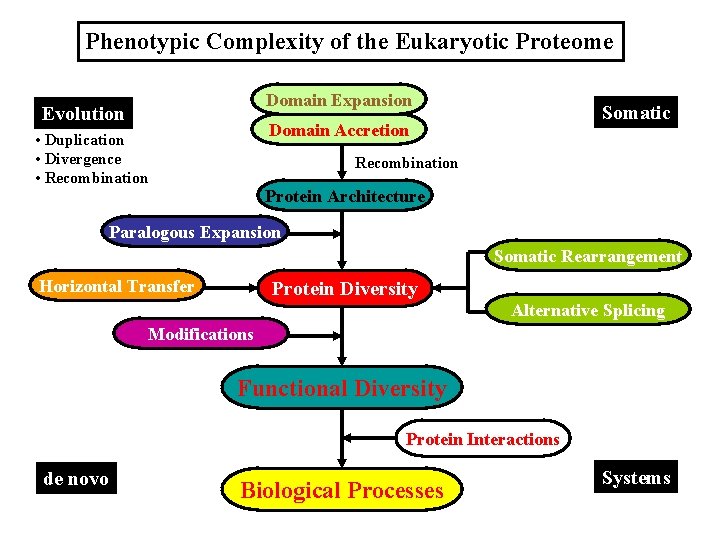
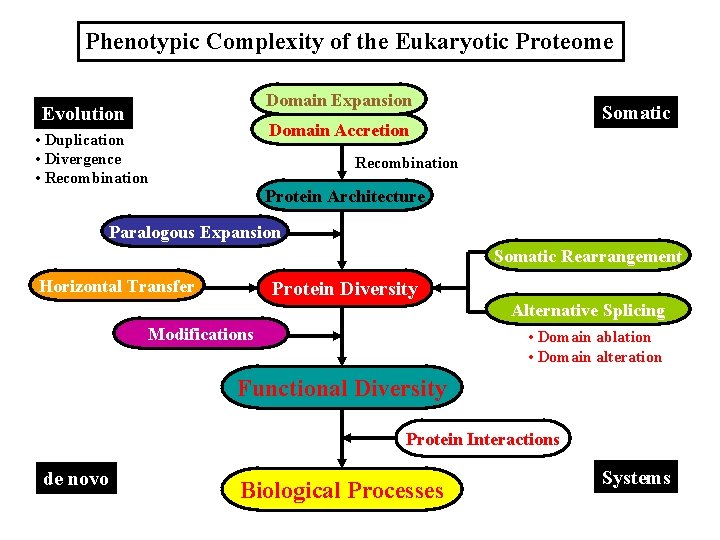
Phenotypic Complexity of the Eukaryotic Proteome Domain Expansion Evolution Somatic Domain Accretion • Duplication • Divergence • Recombination Protein Architecture Paralogous Expansion Somatic Rearrangement Horizontal Transfer Protein Diversity Alternative Splicing Modifications Functional Diversity Protein Interactions de novo Biological Processes Systems

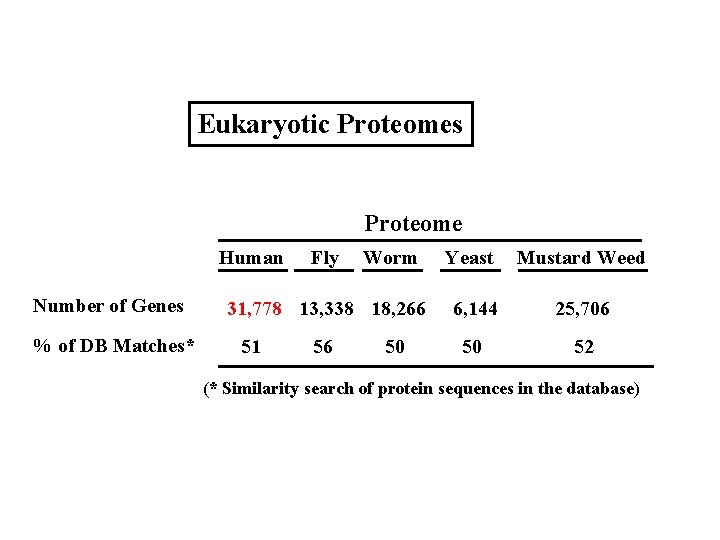
Eukaryotic Proteomes Proteome Human Number of Genes % of DB Matches* Fly Worm 31, 778 13, 338 18, 266 51 56 50 Yeast Mustard Weed 6, 144 25, 706 50 52 (* Similarity search of protein sequences in the database)

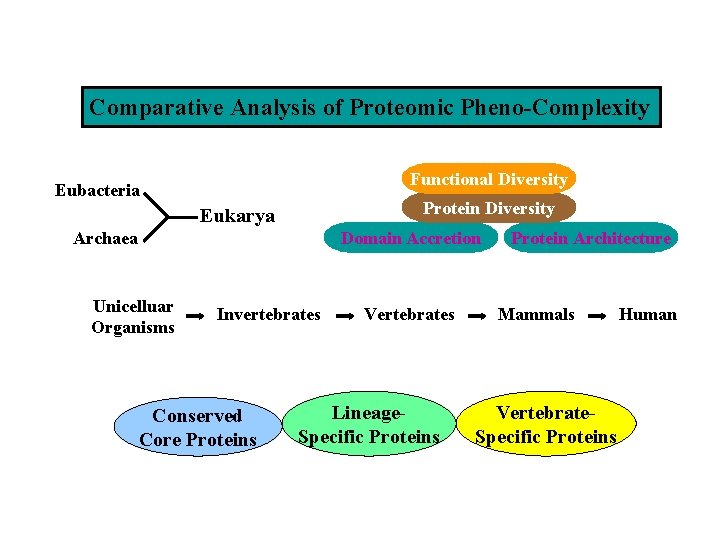
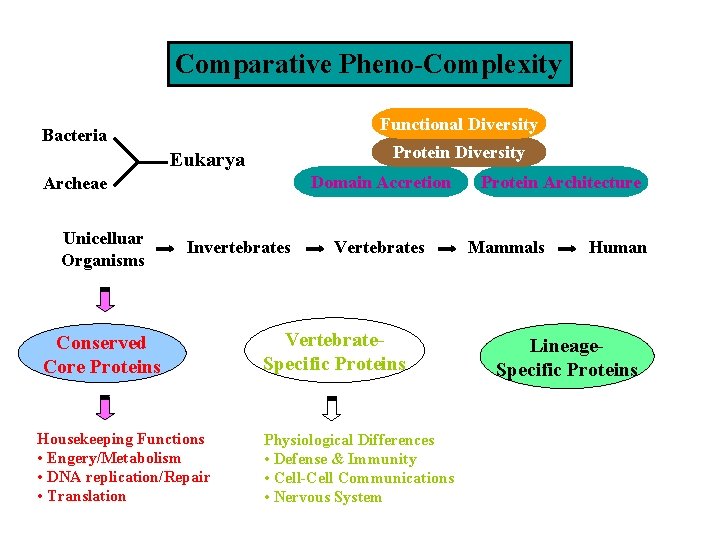
Comparative Analysis of Proteomic Pheno-Complexity Functional Diversity Eubacteria Protein Diversity Eukarya Domain Accretion Archaea Unicelluar Organisms Invertebrates Conserved Core Proteins Vertebrates Lineage. Specific Proteins Protein Architecture Mammals Vertebrate. Specific Proteins Human

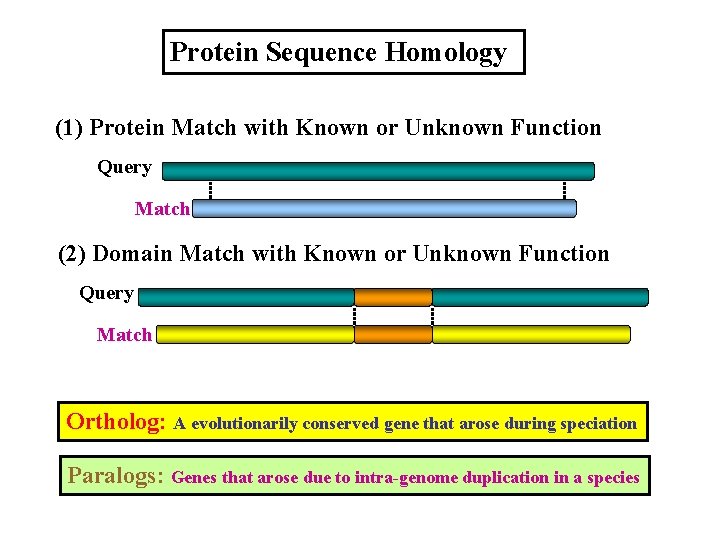
Protein Sequence Homology (1) Protein Match with Known or Unknown Function Query Match (2) Domain Match with Known or Unknown Function Query Match Ortholog: A evolutionarily conserved gene that arose during speciation Paralogs: Genes that arose due to intra-genome duplication in a species

Protein Sequence Comparison (I) Homology • > 40 % : Same Function • 25 -40 % : Similar Function • < 25 % : Different Function (II) Distance • Phylogenetic Tree

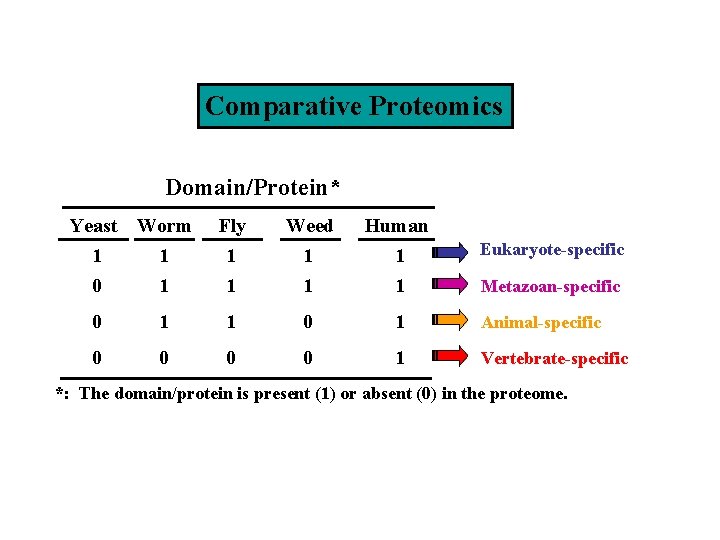
Comparative Proteomics Domain/Protein* Yeast Worm Fly Weed Human 1 0 1 1 1 1 Eukaryote-specific 0 1 1 0 1 Animal-specific 0 0 1 Vertebrate-specific Metazoan-specific *: The domain/protein is present (1) or absent (0) in the proteome.

Eukaryotic Proteomes Shared with Humans Human 61% 43% Fly Worm 46% Yeast

Conserved Core Groups in Eukaryotes Human (3, 109 Proteins) Conserved Fly Yeast Core Proteins in (1, 445 Proteins) (1, 441 Proteins) 1, 308 Groups Worm (1, 503 Proteins)

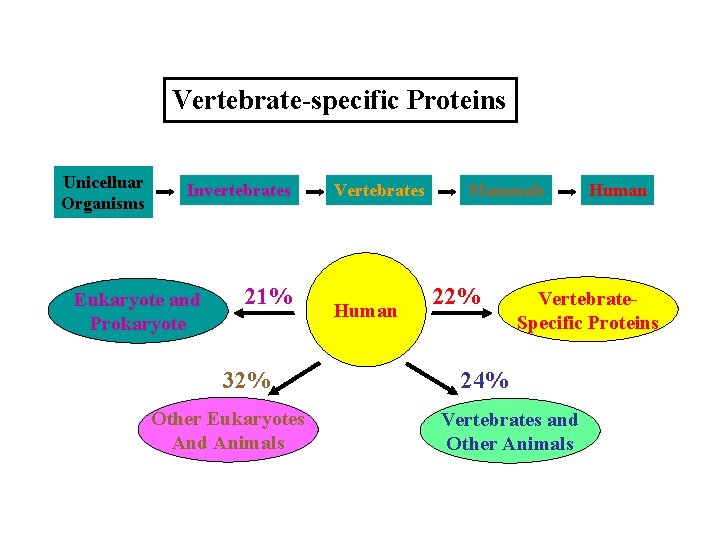
Vertebrate-specific Proteins Unicelluar Organisms Invertebrates Eukaryote and Prokaryote 21% 32% Other Eukaryotes And Animals Vertebrates Human Mammals 22% Human Vertebrate. Specific Proteins 24% Vertebrates and Other Animals

Comparative Pheno-Complexity Functional Diversity Bacteria Protein Diversity Eukarya Domain Accretion Archeae Unicelluar Organisms Invertebrates Conserved Core Proteins Housekeeping Functions • Engery/Metabolism • DNA replication/Repair • Translation Vertebrates Vertebrate. Specific Proteins Physiological Differences • Defense & Immunity • Cell-Cell Communications • Nervous System Protein Architecture Mammals Human Lineage. Specific Proteins

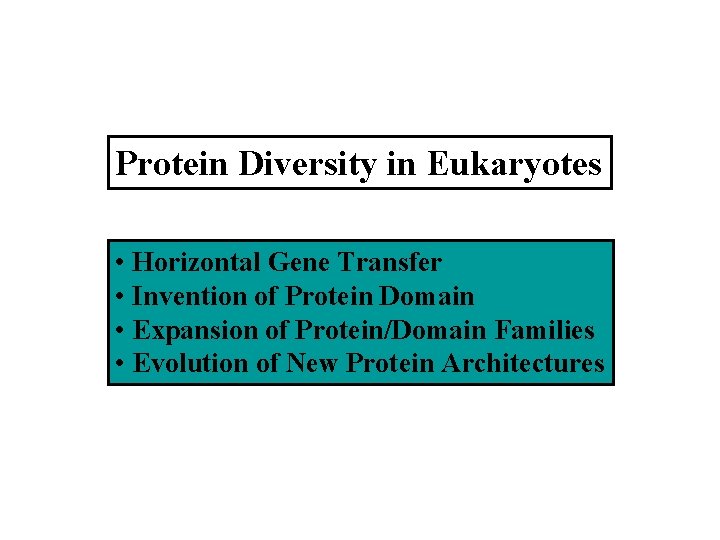
Protein Diversity in Eukaryotes • Horizontal Gene Transfer • Invention of Protein Domain • Expansion of Protein/Domain Families • Evolution of New Protein Architectures

Lateral Gene Transfer Bacteria 223 Genes • Hydrolase • Oxidoreductase • Dehydrogenase • Monoamine Oxidase • Transporter Human • Lineage Specific • Intron Acquisition

Comparative Pheno-Complexity Functional Diversity Bacteria Protein Diversity Eukarya Domain Accretion Archeae Unicelluar Organisms Invertebrates Conserved Core Proteins Housekeeping Functions • Engery/Metabolism • DNA replication/Repair • Translation Vertebrates Vertebrate. Specific Proteins Physiological Differences • Defense & Immunity • Cell-Cell Communications • Nervous System Protein Architecture Mammals Human Lineage. Specific Proteins

Protein Function Assignment 12 Function Categories (Gene Ontology Project) 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. Cellular Processes Metabolism DNA Replication/Modification Transcription/Translation Intracellular Signaling Cell-Cell Communication Protein Folding/Degradation Transport Multifunctional Proteins Cytoskeletal/Structural Defense and Immunity Miscellaneous Function

Classification of Proteome (1) Functional Categories (2) Evolutionary Conservation (3) Structural Classification Cellular Function Protein Sequence Functional Annotation Domain/Motif Databases PRINTS, Prosite, Pfam, Prosite Profile ~50% of Eukaryotes

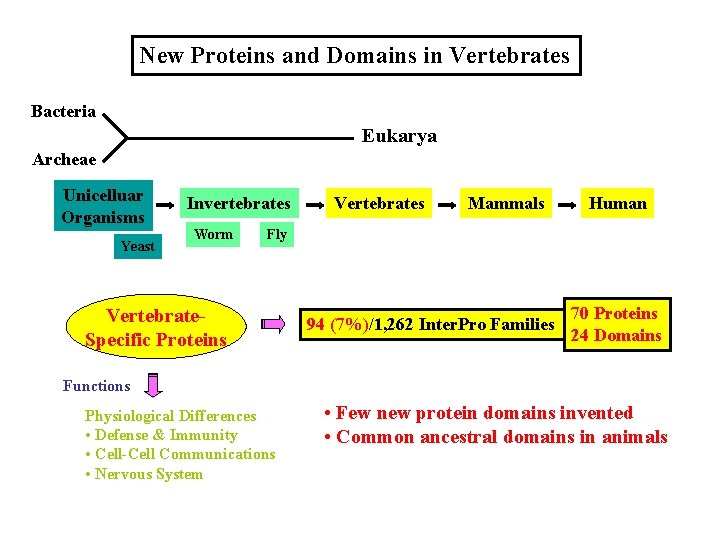
New Proteins and Domains in Vertebrates Bacteria Eukarya Archeae Unicelluar Organisms Yeast Invertebrates Worm Vertebrates Mammals Human Fly Vertebrate. Specific Proteins 94 (7%)/1, 262 Inter. Pro Families 70 Proteins 24 Domains Functions Physiological Differences • Defense & Immunity • Cell-Cell Communications • Nervous System • Few new protein domains invented • Common ancestral domains in animals

Protein Domain • An evolutionary unit • The coding sequence can be duplicated and/or recombined • ~100 to 250 residues • In small proteins or parts of large ones in a domain family • Descending from a common ancestor • Duplication: to give arise one or more domains • Divergence: to generate modified proteins by mutations or In/Del • Recombination: to produce new domain arrangements

Protein Domain Architecture (1) Single-domain Protein (II) Multi-domain Protein Domain A B C D • Prokaryotic Proteome: 2/3 proteins are > 2 domains • Eukaryotic Proteome: 4/5 proteins are multi-domain

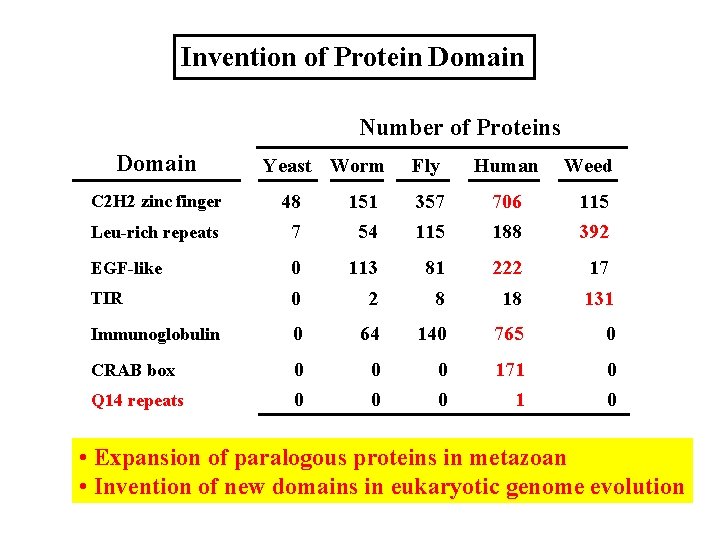
Invention of Protein Domain Number of Proteins Domain Yeast Worm Fly Human Weed 48 7 151 54 357 115 706 188 115 392 EGF-like 0 113 81 222 17 TIR 0 2 8 18 131 Immunoglobulin 0 64 140 765 0 CRAB box 0 0 0 171 1 0 0 C 2 H 2 zinc finger Leu-rich repeats Q 14 repeats • Expansion of paralogous proteins in metazoan • Invention of new domains in eukaryotic genome evolution

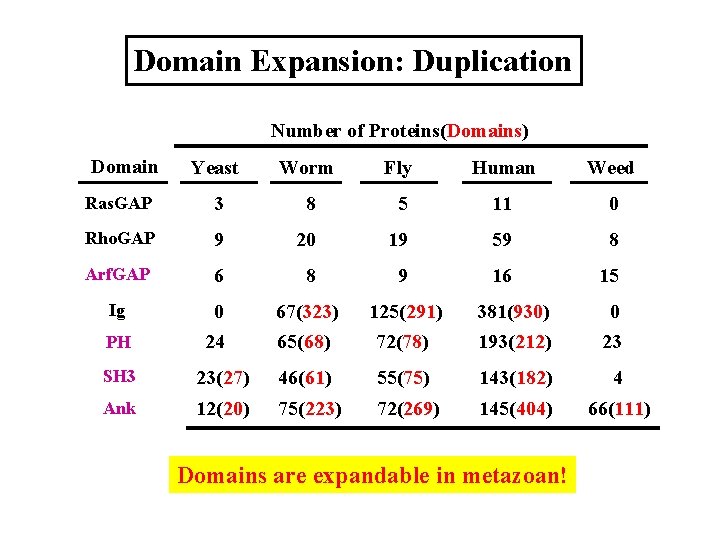
Domain Expansion: Duplication Number of Proteins(Domains) Domain Yeast Worm Fly Human Weed Ras. GAP 3 8 5 11 0 Rho. GAP 9 20 19 59 8 Arf. GAP 6 8 9 16 15 Ig 0 24 67(323) 65(68) 125(291) 72(78) 381(930) 193(212) 0 23 PH SH 3 23(27) 46(61) 55(75) 143(182) 4 Ank 12(20) 75(223) 72(269) 145(404) 66(111) Domains are expandable in metazoan!

Rosetta Stone Similarity Search of Protein Databases Function 1 Protein A Protein B Function 2 Protein X Functions 1 and 2 due to domain recombination

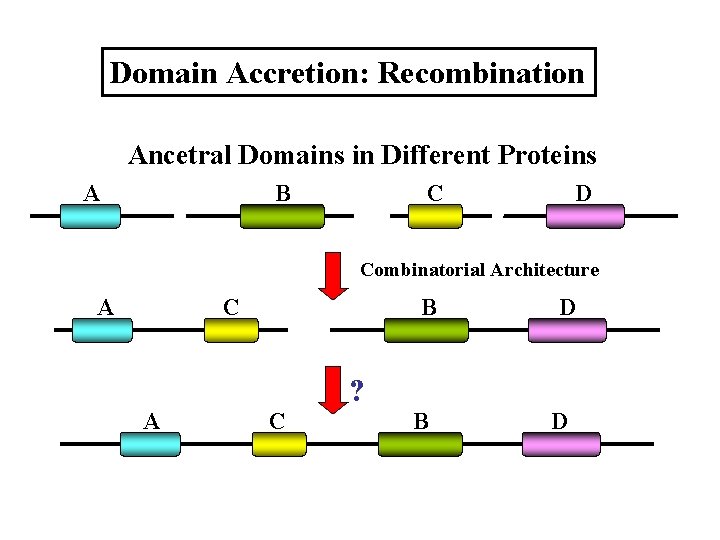
Domain Accretion: Recombination Ancetral Domains in Different Proteins A B C D Combinatorial Architecture A C A B C ? B D D

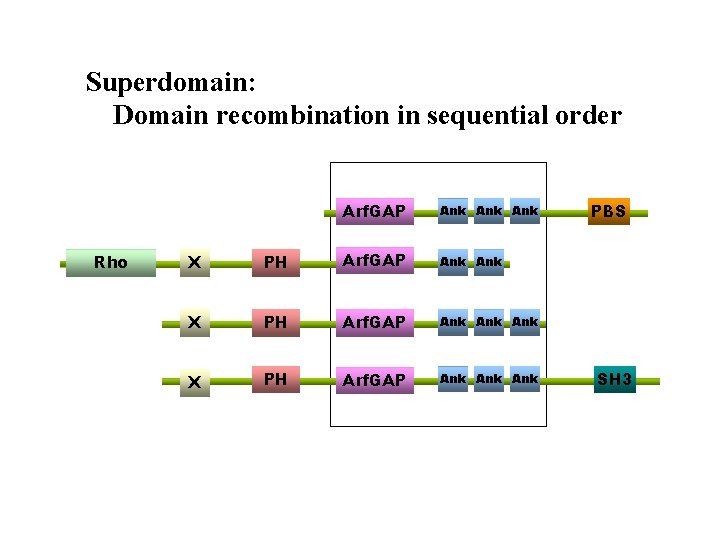
Superdomain: Domain recombination in sequential order Rho Arf. GAP Ank Ank X PH Arf. GAP Ank Ank PBS SH 3

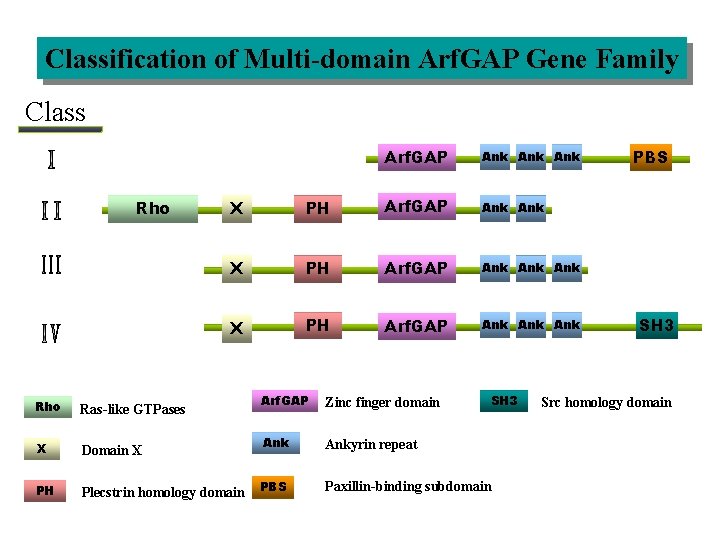
Classification of Multi-domain Arf. GAP Gene Family Class Rho Arf. GAP Ank Ank X PH Arf. GAP Ank Ank Rho Ras-like GTPases X Domain X PH Plecstrin homology domain Arf. GAP Zinc finger domain Ankyrin repeat PBS SH 3 Paxillin-binding subdomain PBS SH 3 Src homology domain

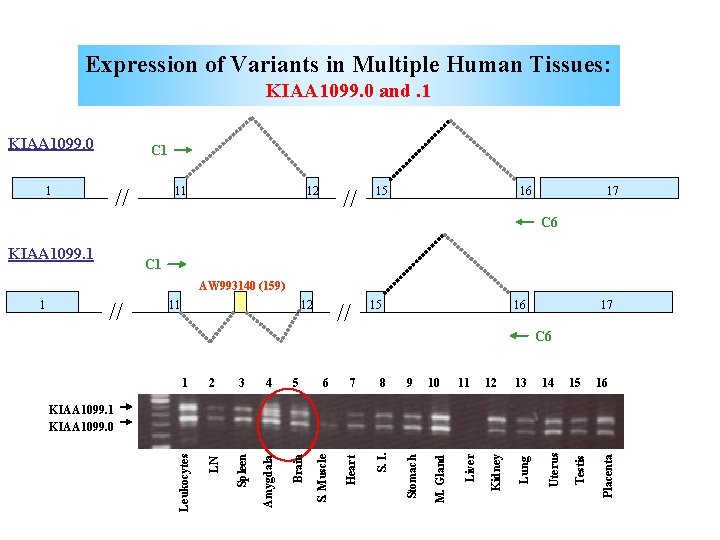
Expression of Variants in Multiple Human Tissues: KIAA 1099. 0 and. 1 KIAA 1099. 0 C 1 // 1 11 12 // 15 16 17 C 6 KIAA 1099. 1 C 1 AW 993140 (159) // 11 12 // 15 16 17 5 Spleen Amygdala Brain 6 7 8 9 Stomach 4 S. I. 3 Heart 2 S. Muscle 1 LN C 6 Leukocytes 10 11 12 13 14 15 16 Placenta Testis Uterus Lung Kidney Liver KIAA 1099. 1 KIAA 1099. 0 M. Gland 1

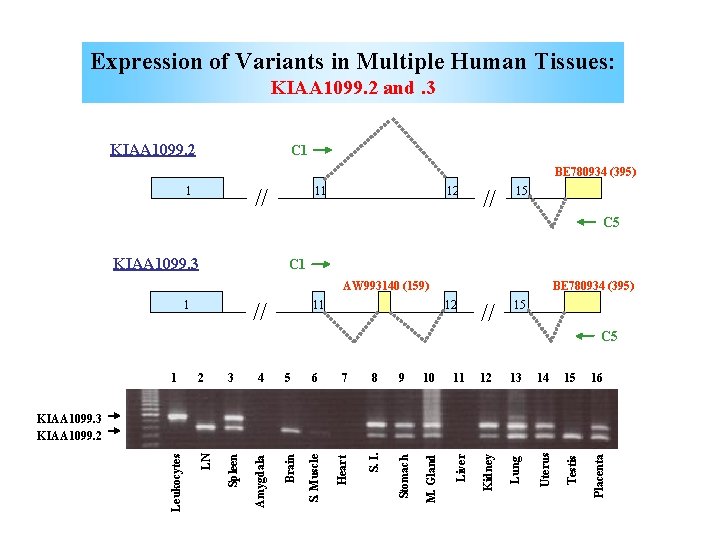
Expression of Variants in Multiple Human Tissues: KIAA 1099. 2 and. 3 KIAA 1099. 2 C 1 BE 780934 (395) // 1 11 12 // 15 C 5 KIAA 1099. 3 C 1 AW 993140 (159) // 1 11 BE 780934 (395) 12 // 15 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 Leukocytes LN Spleen Amygdala Brain S. Muscle Heart S. I. Stomach M. Gland Liver Kidney Lung Uterus Testis Placenta C 5 KIAA 1099. 3 KIAA 1099. 2

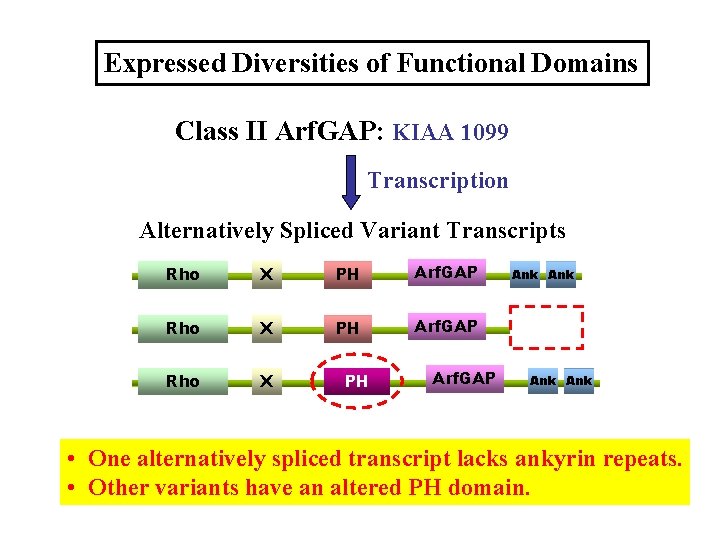
Expressed Diversities of Functional Domains Class II Arf. GAP: KIAA 1099 Transcription Alternatively Spliced Variant Transcripts Rho X PH Arf. GAP Ank Ank • One alternatively spliced transcript lacks ankyrin repeats. • Other variants have an altered PH domain.

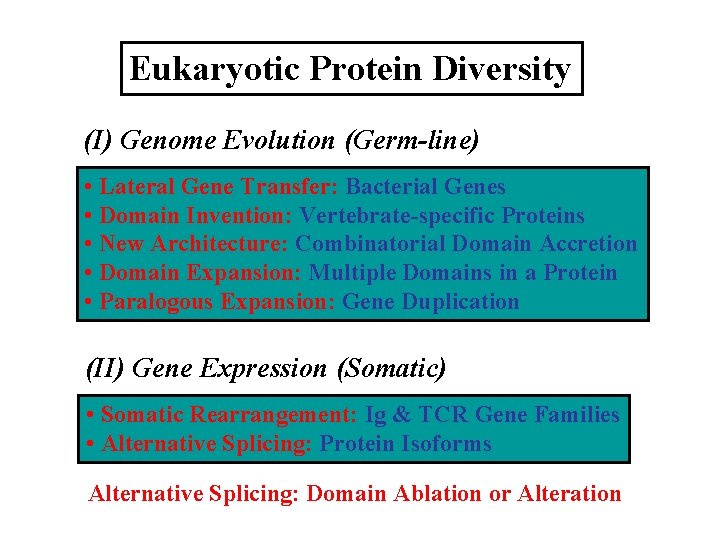
Eukaryotic Protein Diversity (I) Genome Evolution (Germ-line) • Lateral Gene Transfer: Bacterial Genes • Domain Invention: Vertebrate-specific Proteins • New Architecture: Combinatorial Domain Accretion • Domain Expansion: Multiple Domains in a Protein • Paralogous Expansion: Gene Duplication (II) Gene Expression (Somatic) • Somatic Rearrangement: Ig & TCR Gene Families • Alternative Splicing: Protein Isoforms Alternative Splicing: Domain Ablation or Alteration

Phenotypic Complexity of the Eukaryotic Proteome Domain Expansion Evolution Somatic Domain Accretion • Duplication • Divergence • Recombination Protein Architecture Paralogous Expansion Somatic Rearrangement Horizontal Transfer Protein Diversity Modifications Alternative Splicing • Domain ablation • Domain alteration Functional Diversity Protein Interactions de novo Biological Processes Systems

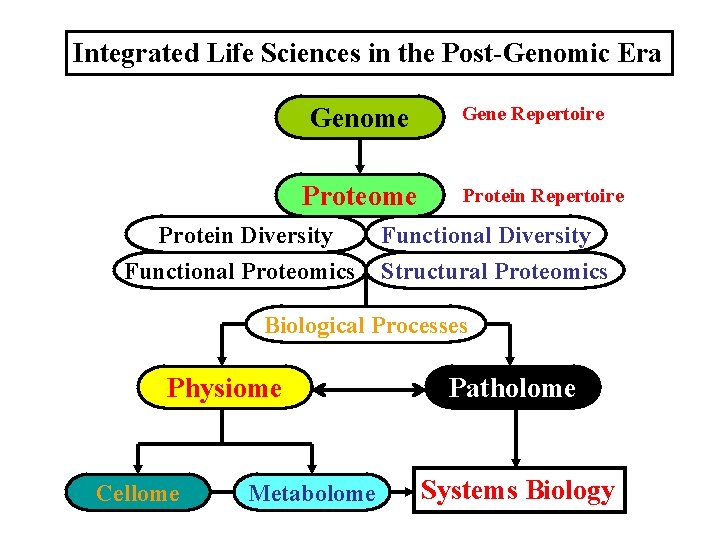
Integrated Life Sciences in the Post-Genomic Era Genome Protein Diversity Functional Proteomics Gene Repertoire Protein Repertoire Functional Diversity Structural Proteomics Biological Processes Physiome Cellome Metabolome Patholome Systems Biology