Applications of molecular simulation in materials science and




- Slides: 7

Applications of molecular simulation in materials science and biology Rebecca Notman Department of Chemistry and Centre for Scientific Computing, University of Warwick

Molecular Dynamics Simulations l Compute the trajectory of a set of interacting particles in a system over time. l Why is it useful? l – To gain a molecular-level view of a system; – To help interpret experiments by linking molecular structure to bulk properties or function; – To act as a bridge between theory and experiment; – To explore extreme conditions. Application of molecular simulation to problems in materials science, pharmaceutical science and biomedicine.

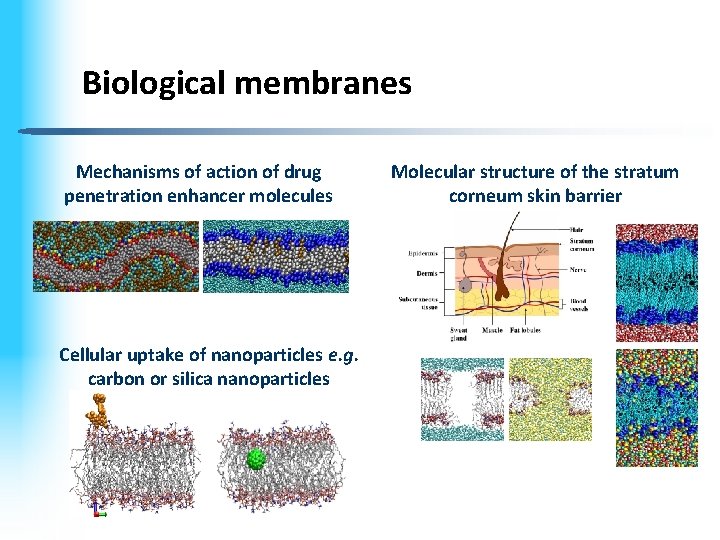
Biological membranes Mechanisms of action of drug penetration enhancer molecules Cellular uptake of nanoparticles e. g. carbon or silica nanoparticles Molecular structure of the stratum corneum skin barrier

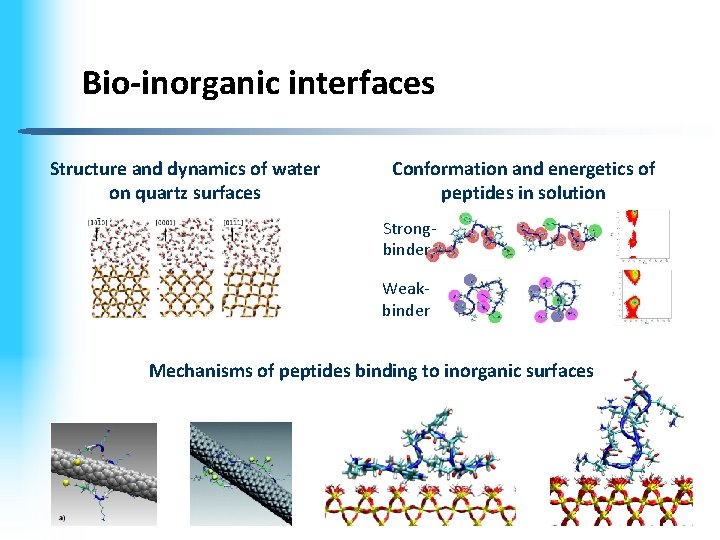
Bio-inorganic interfaces Structure and dynamics of water on quartz surfaces Conformation and energetics of peptides in solution Strongbinder Weakbinder Mechanisms of peptides binding to inorganic surfaces

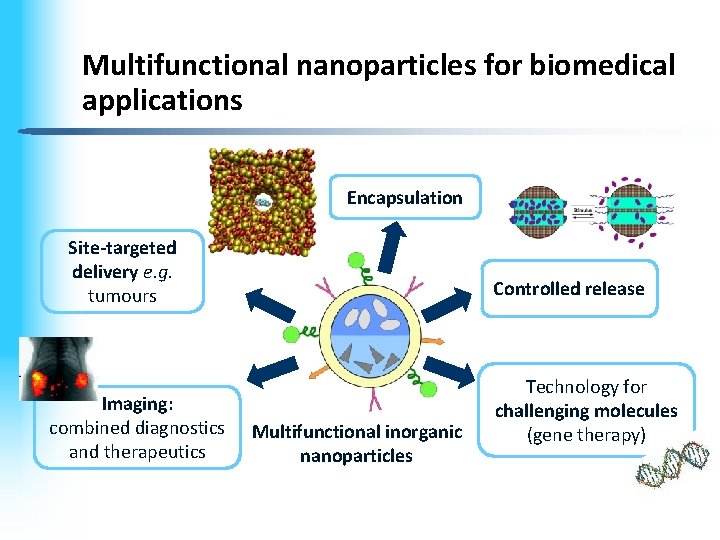
Multifunctional nanoparticles for biomedical applications Encapsulation Site-targeted delivery e. g. tumours Imaging: combined diagnostics and therapeutics Controlled release Multifunctional inorganic nanoparticles Technology for challenging molecules (gene therapy)

Acknowledgements l l Biological membranes: – Jamshed Anwar (Bradford) – Wim Briels and Wouter den Otter (Twente) – Massimo Noro (Unilever R&D) Bio-inorganic interfaces: – Tiffany Walsh (Warwick) – Mehmet Sarikaya Group (Washington)

References l l Biological membranes: – Notman, R. , Noro, M. G. , O’Malley, B. and Anwar, J. , Molecular basis for dimethylsulfoxide (DMSO) action on lipid membranes, J. Am. Chem. Soc. , 2006, 128: 13982. – Notman, R. , Noro, M. G. and Anwar, J. , Interaction of oleic acid with dipalmitoylphosphatidylcholine (DPPC) bilayers simulated by molecular dynamics, J. Phys. Chem. B. , 2007, 111: 12748. – Notman, R. , den Otter, W. K. , Noro, M. G. , Briels, W. J. and Anwar, J. , The permeability enhancing mechanism of DMSO in ceramide bilayers simulated by molecular dynamics, Biophys. J. , 2007, 93: 2056. – Notman, R. , Anwar J. , Briels, W. K. , Noro, M. G. and den Otter, W. K. , Simulations of skin barrier function: Free energies of hydrophobic and hydrophilic transmembrane pores in ceramide bilayers, Biophys. J. , 2008, 95: 4763. Bio-inorganic interfaces: – Notman, R. and Walsh, T. R. , Molecular dynamics studies of the interactions of water and amino acid analogues with quartz surfaces, Langmuir, 2009, 3: 1638. – Friling, S. R. , Notman, R. and Walsh, T. R. , Probing diameter-selective solubilisation of carbon nanotubes by reversible cyclic peptides using molecular dynamics simulations, Nanoscale, 2010, in press. – Oren, E. E. , Notman, R. , Kim, I. W. , Evans, J. S. , Walsh, T. R. , Samudrala, R. , Tamerler, C. and Sarikaya, M. , Probing the molecular mechanisms of solid-binding peptides, Submitted, 2010. – Notman, R. and Walsh, T. R. Solution studies of strong and weak quartz-binding peptides using replica exchange molecular dynamics, Submitted, 2010.