Application of Bioinformatics Proteomics and Genomics ABPG Introns

Application of Bioinformatics, Proteomics, and Genomics (ABPG) Introns, Spcicing, and Alternative splicing
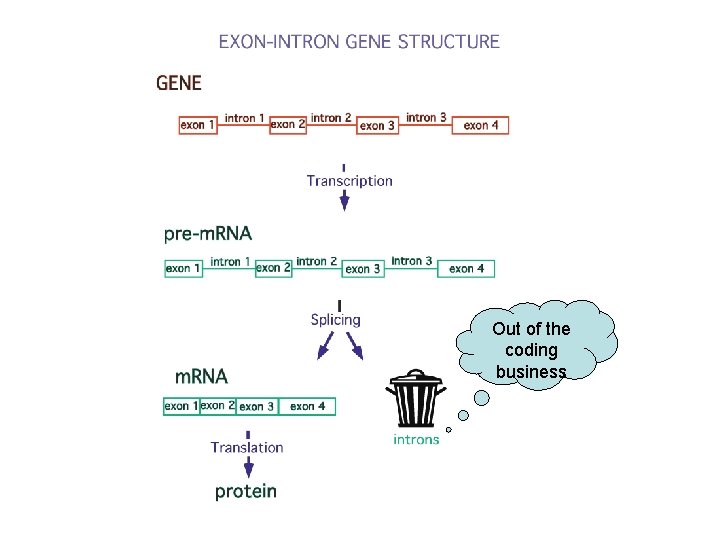
Out of the coding business
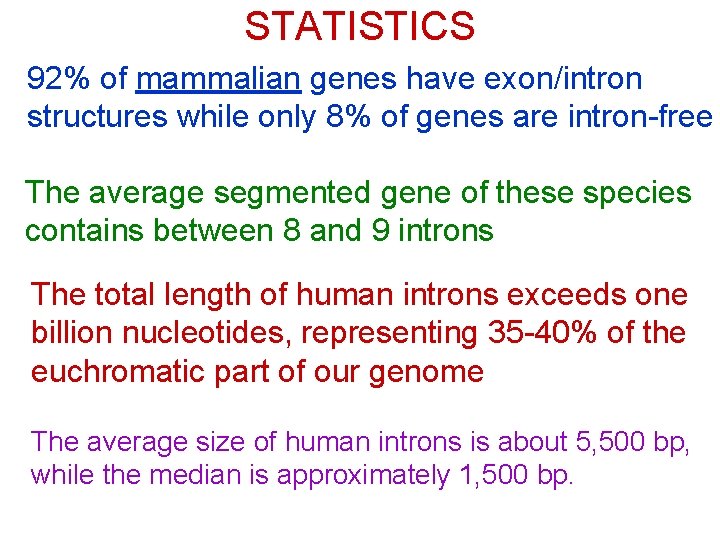
STATISTICS 92% of mammalian genes have exon/intron structures while only 8% of genes are intron-free The average segmented gene of these species contains between 8 and 9 introns The total length of human introns exceeds one billion nucleotides, representing 35 -40% of the euchromatic part of our genome The average size of human introns is about 5, 500 bp, while the median is approximately 1, 500 bp.
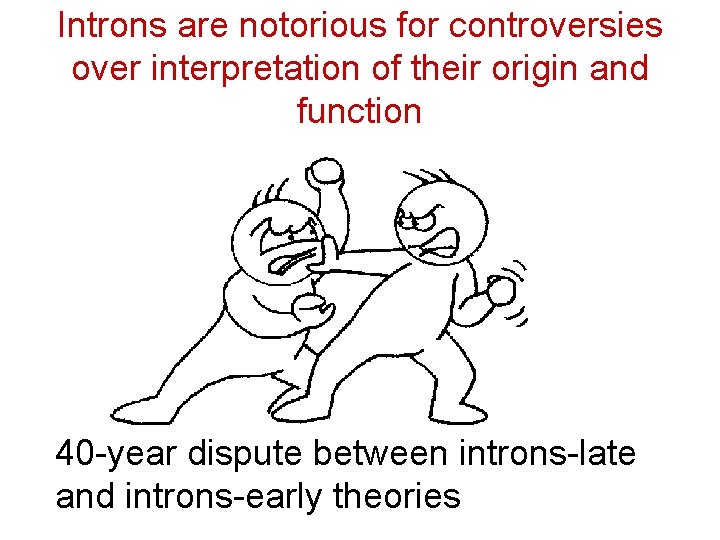
Introns are notorious for controversies over interpretation of their origin and function 40 -year dispute between introns-late and introns-early theories

What do we know about introns? • Introns are the ubiquitous genomic elements of eukaryotes whose role is still poorly understood and appreciated.
Tree of life
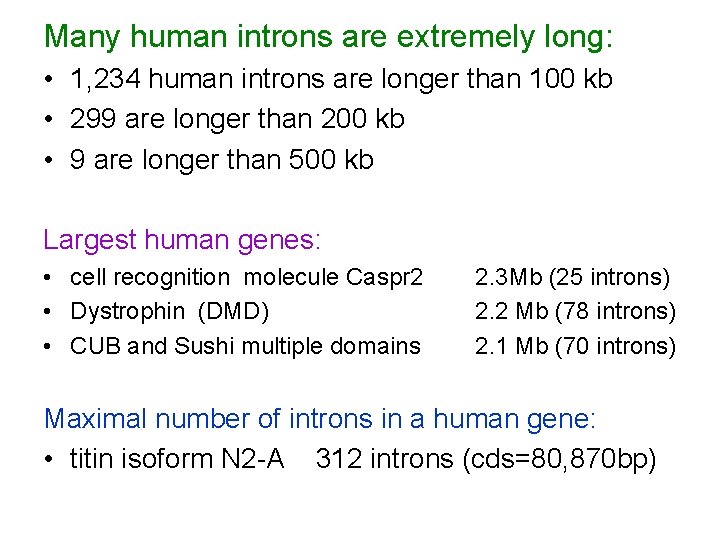
Many human introns are extremely long: • 1, 234 human introns are longer than 100 kb • 299 are longer than 200 kb • 9 are longer than 500 kb Largest human genes: • cell recognition molecule Caspr 2 • Dystrophin (DMD) • CUB and Sushi multiple domains 2. 3 Mb (25 introns) 2. 2 Mb (78 introns) 2. 1 Mb (70 introns) Maximal number of introns in a human gene: • titin isoform N 2 -A 312 introns (cds=80, 870 bp)
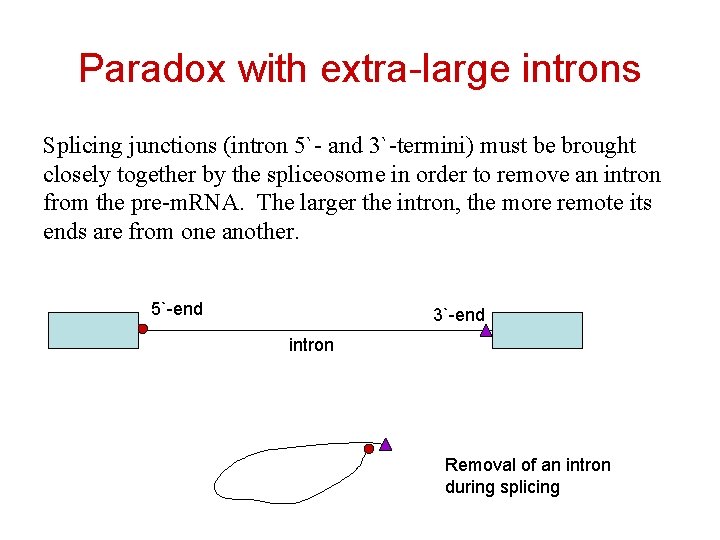
Paradox with extra-large introns Splicing junctions (intron 5`- and 3`-termini) must be brought closely together by the spliceosome in order to remove an intron from the pre-m. RNA. The larger the intron, the more remote its ends are from one another. 5`-end 3`-end intron Removal of an intron during splicing
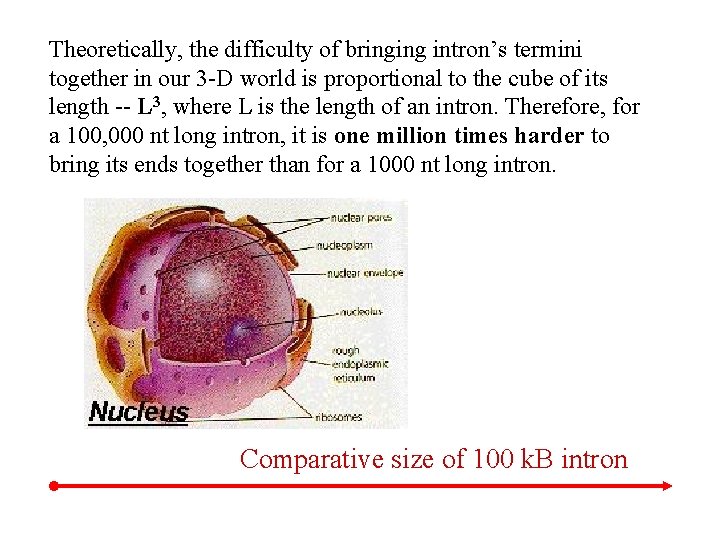
Theoretically, the difficulty of bringing intron’s termini together in our 3 -D world is proportional to the cube of its length -- L 3, where L is the length of an intron. Therefore, for a 100, 000 nt long intron, it is one million times harder to bring its ends together than for a 1000 nt long intron. Comparative size of 100 k. B intron
The enormous intron size in mammals creates several drawbacks, such as: 1) considerable waste of energy during gene expression, which is “unwisely” spent on polymerizing extra-long intronic segments of pre-m. RNA molecules; 2) delay in obtaining protein products (on average it takes about 45 min for RNA polymerase II to transcribe a 100, 000 bp intron); 3) potential errors in normal splicing, since long introns contain numerous false splice sites (so-called pseudoexons). Some benefits must be associated with introns to compensate for these disadvantages. Different constructive roles for introns are described in two reviews: Fedorova L. , Fedorov A. Introns in gene evolution. Genetica 2003, 118: 123 -131. Fedorova L. , Fedorov A. Puzzles of the human genome: why do we need our introns? Current Genomics 2005, Vol. 6, 589 -595.

Intron functions 1) sources of non-coding RNA 2) carriers of transcription regulatory elements 3) actors in alternative and trans-splicing 4) enhancers of meiotic crossing over within coding sequences 5) substrates for exon shuffling 6) signals for m. RNA export from the nucleus and nonsense-mediated decay

• Removal of nuclear (spliceosomal) introns is extremely complex process which requires up to 250 proteins and several small non-coding RNAs (U 1, U 2, U 4, U 5, U 6) http: //www. ncbi. nlm. nih. gov/books/bv. fcgi? rid=mcb. figgrp. 2890 • Video http: //vcell. ndsu. edu/animations/mrnasplicing/movie-flash. htm
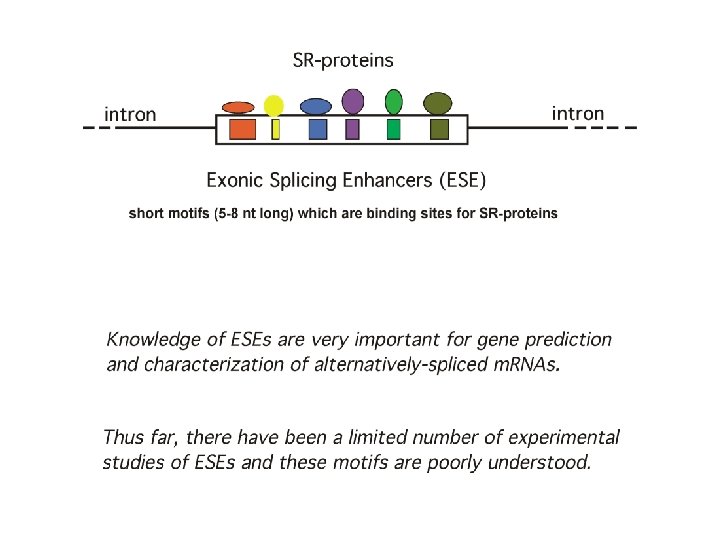
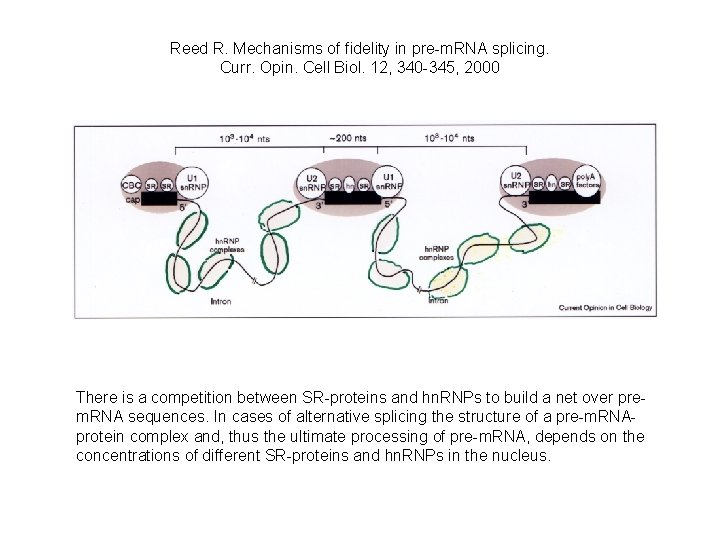
Reed R. Mechanisms of fidelity in pre-m. RNA splicing. Curr. Opin. Cell Biol. 12, 340 -345, 2000 There is a competition between SR-proteins and hn. RNPs to build a net over prem. RNA sequences. In cases of alternative splicing the structure of a pre-m. RNAprotein complex and, thus the ultimate processing of pre-m. RNA, depends on the concentrations of different SR-proteins and hn. RNPs in the nucleus.
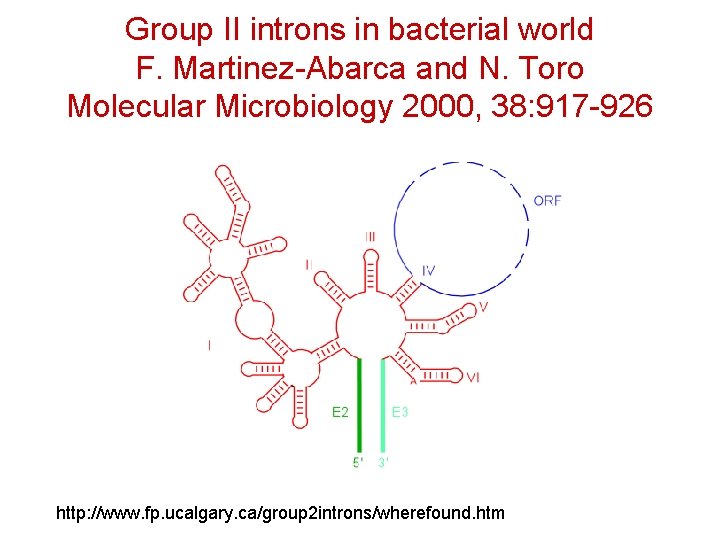
Group II introns in bacterial world F. Martinez-Abarca and N. Toro Molecular Microbiology 2000, 38: 917 -926 http: //www. fp. ucalgary. ca/group 2 introns/wherefound. htm
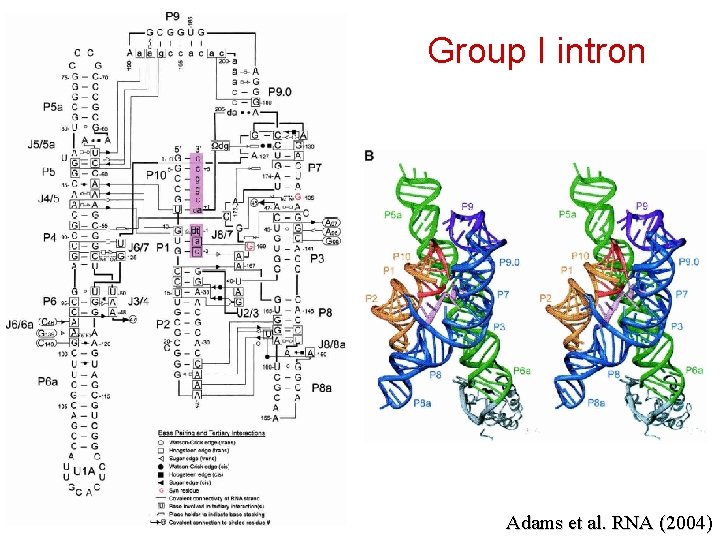
Group I intron Adams et al. RNA (2004)
Evolutionarily distant species share only a portion of common introns Animals and plants have no more than 50% of common intron positions in orthologous genes Fedorov et al. PNAS 2002, 99: 16128 Rogozin et al. Curr Biol 2003, 13: 1512
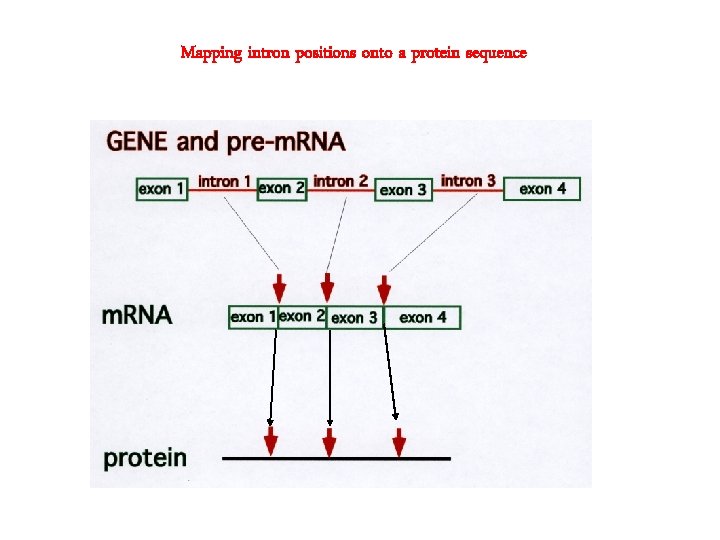
Mapping intron positions onto a protein sequence
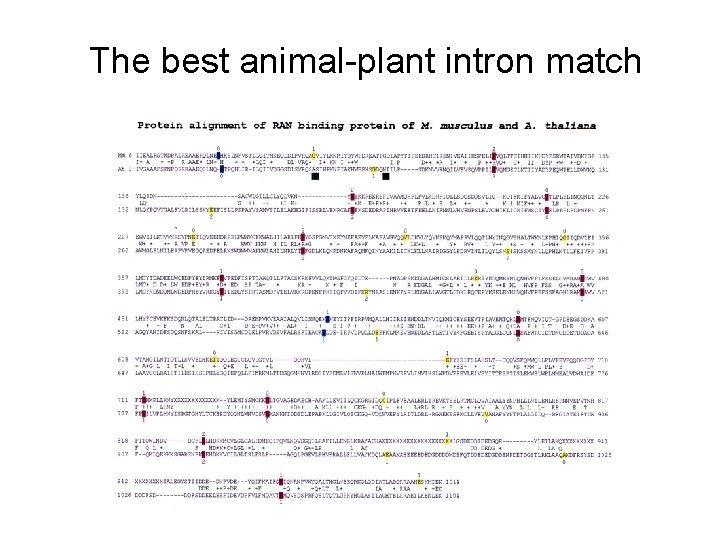
The best animal-plant intron match

Intron loss and gain during evolution
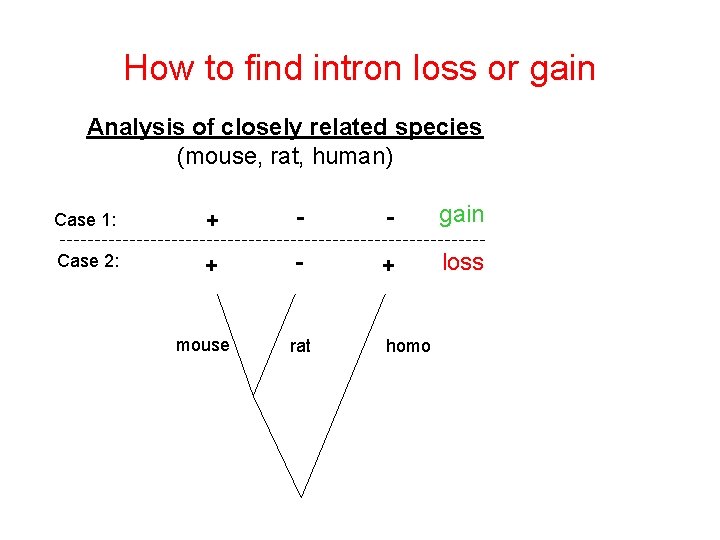
How to find intron loss or gain Analysis of closely related species (mouse, rat, human) Case 1: + - - gain Case 2: + - + loss rat homo mouse
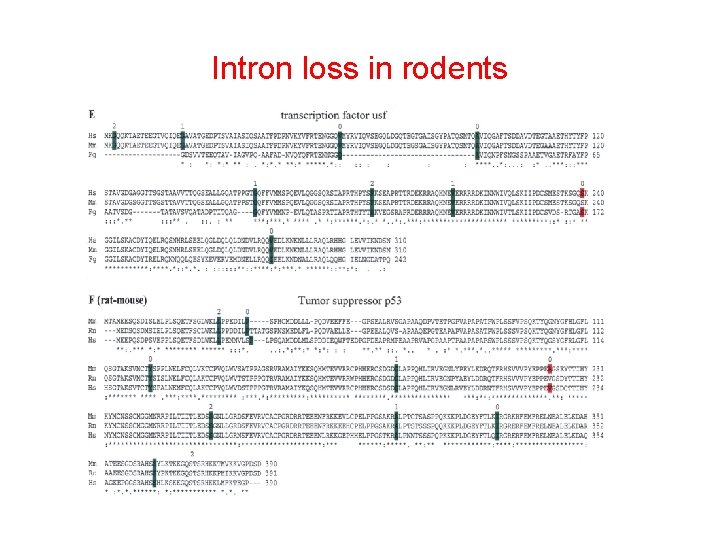
Intron loss in rodents

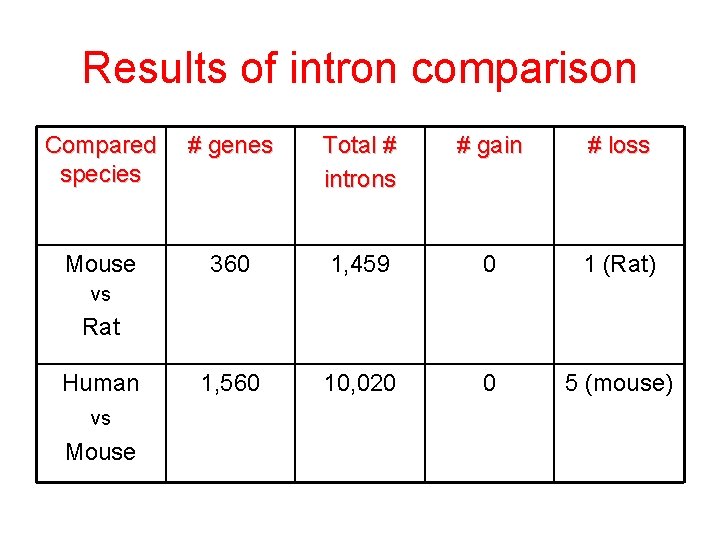
Results of intron comparison Compared species # genes Total # introns # gain # loss Mouse 360 1, 459 0 1 (Rat) 1, 560 10, 020 0 5 (mouse) vs Rat Human vs Mouse
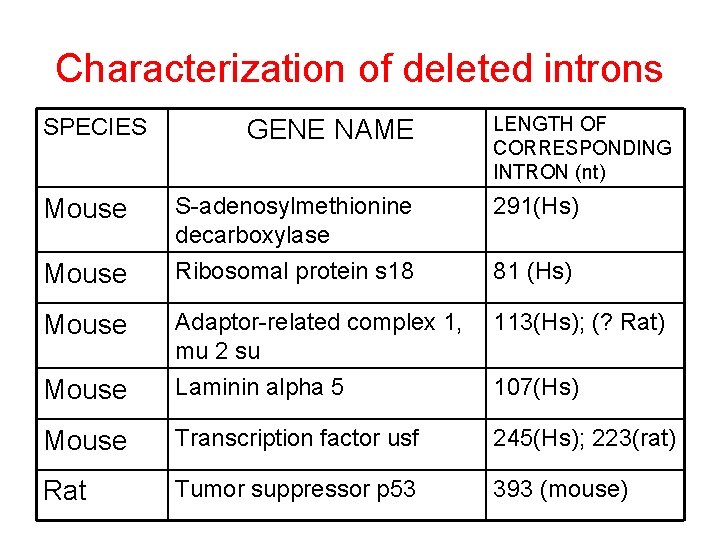
Characterization of deleted introns SPECIES GENE NAME LENGTH OF CORRESPONDING INTRON (nt) S-adenosylmethionine decarboxylase Ribosomal protein s 18 291(Hs) 113(Hs); (? Rat) Mouse Adaptor-related complex 1, mu 2 su Laminin alpha 5 Mouse Transcription factor usf 245(Hs); 223(rat) Rat Tumor suppressor p 53 393 (mouse) Mouse 81 (Hs) 107(Hs)
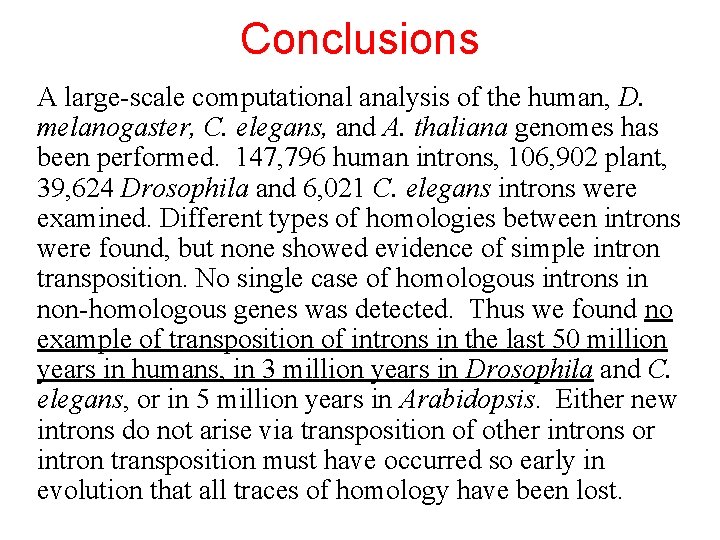
Conclusions A large-scale computational analysis of the human, D. melanogaster, C. elegans, and A. thaliana genomes has been performed. 147, 796 human introns, 106, 902 plant, 39, 624 Drosophila and 6, 021 C. elegans introns were examined. Different types of homologies between introns were found, but none showed evidence of simple intron transposition. No single case of homologous introns in non-homologous genes was detected. Thus we found no example of transposition of introns in the last 50 million years in humans, in 3 million years in Drosophila and C. elegans, or in 5 million years in Arabidopsis. Either new introns do not arise via transposition of other introns or intron transposition must have occurred so early in evolution that all traces of homology have been lost.

Genome Research 2003 For nearly 15 years, it has been widely believed that many introns were recently acquired by the genes of multicellular organisms. However, the mechanism of acquisition has yet to be described for a single animal intron. Here, we report a large-scale computational analysis of the human, Drosophila melanogaster, Caenorhabditis elegans, and Arabidopsis thaliana genomes. We divided 147, 796 human intron sequences into batches of similar lengths and aligned them with each other. Different types of homologies between introns were found, but none showed evidence of simple intron transposition.

Alternative splicing Production of multiple m. RNA isoforms from the same gene often in a tissue-specific or development-stage-specific manner
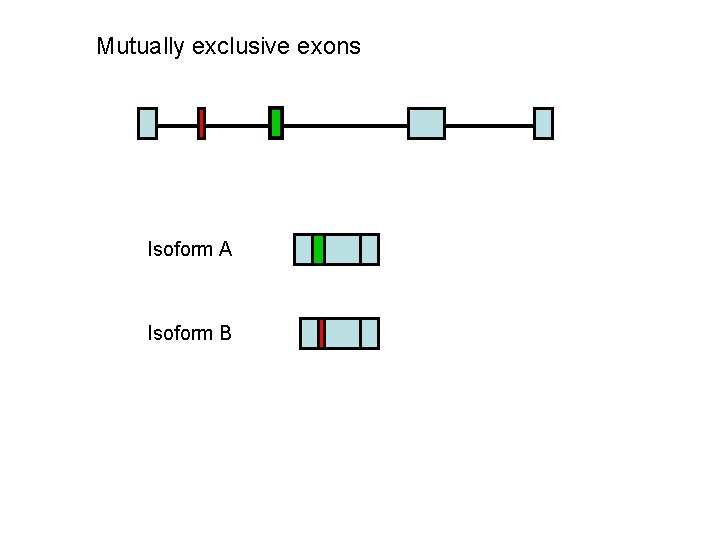
Mutually exclusive exons Isoform A Isoform B
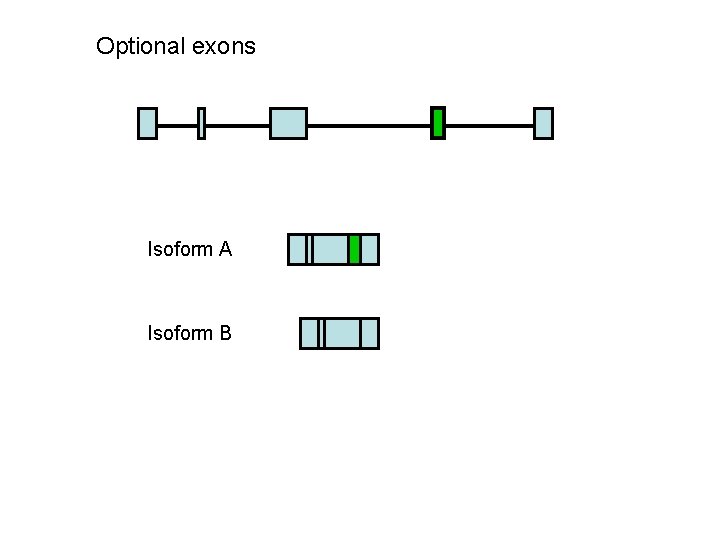
Optional exons Isoform A Isoform B
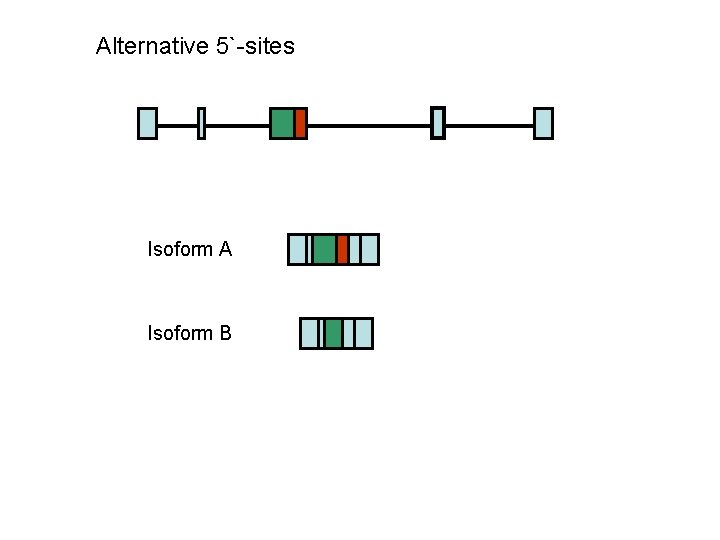
Alternative 5`-sites Isoform A Isoform B
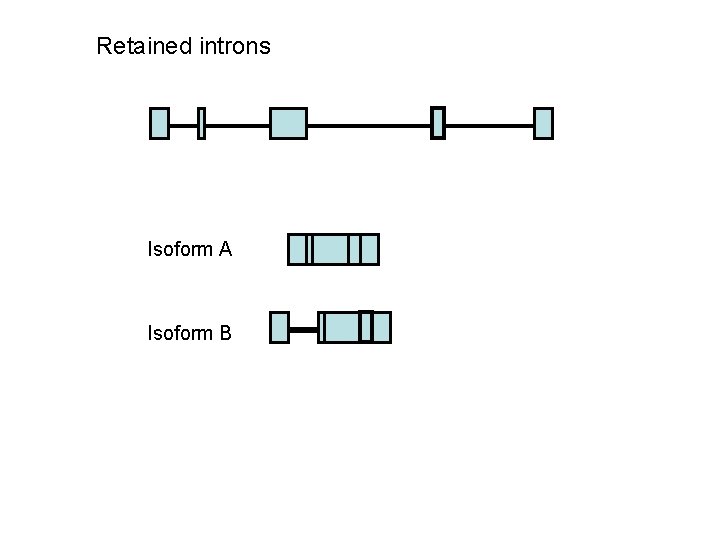
Retained introns Isoform A Isoform B

Why alternative splicing is important ? • Half of human genes express multiple alternative m. RNA isoforms, many of which have important specific functions. • Alternative splicing alone increases the number of different polypeptides in human cells by 2 -3 fold above the number of human genes

Alternative splicing in Drosophila melanogaster The study of sex-determination development in Drosophila involving the alternatively spliced Sex-lethal (Sxl), male-specific-lethal-2 (msl 2), transformer (tra), and doublesex (dsx) genes led to the initial discovery of ESE (reviewed by Cline and Meyer 1996; Mac. Dougall et al. 1995).
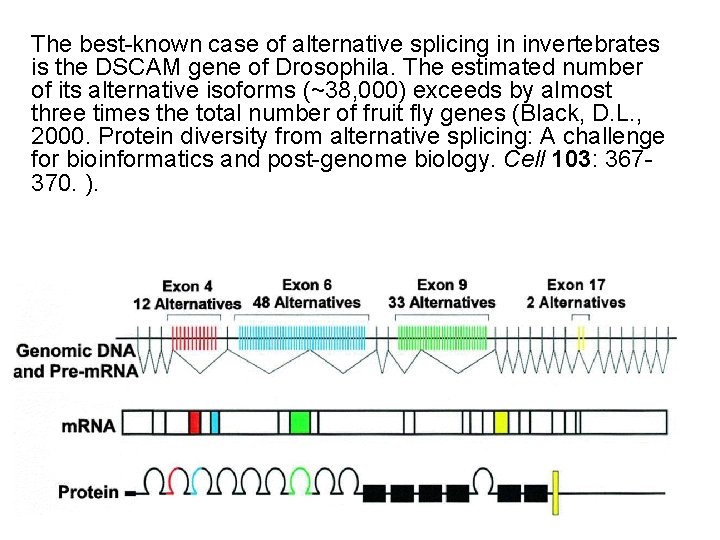
The best-known case of alternative splicing in invertebrates is the DSCAM gene of Drosophila. The estimated number of its alternative isoforms (~38, 000) exceeds by almost three times the total number of fruit fly genes (Black, D. L. , 2000. Protein diversity from alternative splicing: A challenge for bioinformatics and post-genome biology. Cell 103: 367370. ).

Alternative splicing in human • Defects in alternative splicing are associated with several human diseases: 1) frontotemporal dementia with parkinsonism, 2) amyotrophic lateral sclerosis, 3) paraneoplastic neurological disorders, 4) maybe some forms of schizophrenia, • Many types of cancer are linked to the altered patterns of alternative splicing. For instance, alternative isoforms of Bcl-2 family of apoptotic regulators have opposite apoptotic activities; frequently anti-apoptotic isoforms are overexpressed in lymphoma cells. • “Neurexin: three genes and 1001 products” TIG 14: 20 -26, 1998. Missler M. and Sudhof T. C.

Comparative genomics Up to 60% of splicing isoforms are conserved between human and mouse What about plants?

For computational biology the most efficient way to study alternative splicing is analysis of EST database
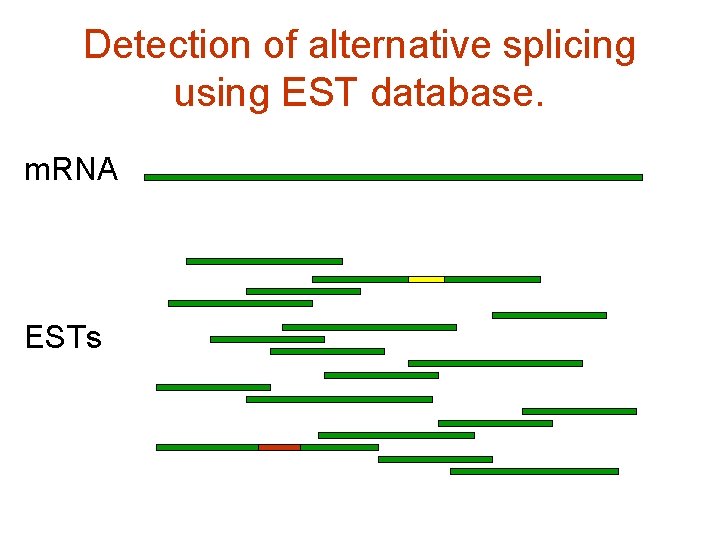
Detection of alternative splicing using EST database. m. RNA ESTs

RNA-Seq Bioinformatics tools http: //en. wikipedia. org/wiki/List_of_RNA-Seq_bioinformatics_tools
This study, along with the following discussion, details the association of thousands of nc. RNAs— sno. RNA, mi. RNA, si. RNA, pi. RNA and long nc. RNA —within human introns. We propose that such an association between human introns and nc. RNAs has a pronounced synergistic effect with important implications for fine-tuning gene expression patterns across the entire genome.
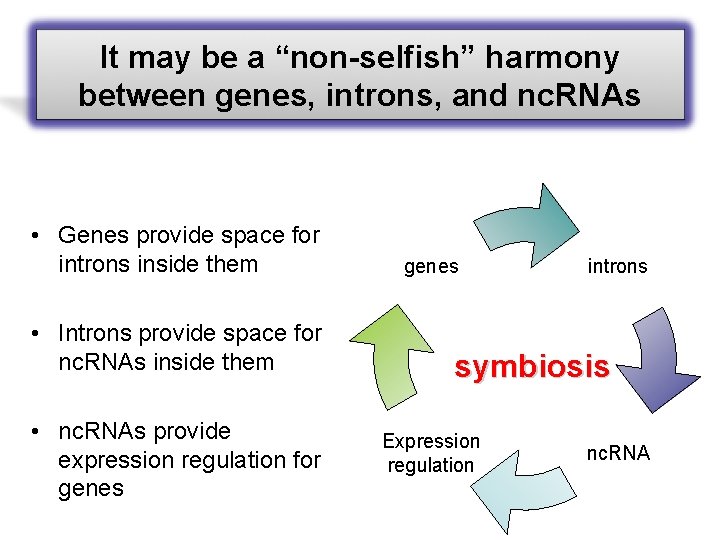
It may be a “non-selfish” harmony between genes, introns, and nc. RNAs • Genes provide space for introns inside them • Introns provide space for nc. RNAs inside them • nc. RNAs provide expression regulation for genes introns symbiosis Expression regulation nc. RNA

HOMEWORK #1 Which intron is it? Does it contain functional elements? > INTRON gtatctctgtatctttatgttgtatcaaacacatgatatttcaca acaagctgaaaagtaggattatgggcaatgccattgtcag cttgttgggcgatatggcaacccactatataatcctctcttaa cagcattgggagtgttgtcaaaaggtttgacagacggttcg gagaactgttgctctaggaggagctgagagttcaagtctct ccatttcccaaaacttttttctcattcacgtggcttgtgtcc tgttccactttgaatatatggctaccccatttgctttcaactgat gtatgatagttttgtcgctttatttcatttttatatattacaatattac caatatctttgtcgttcaccag
OPTIONAL Homework assignment to earn extra credit Why there is a difference in exon-intron structures of rat gaba-receptor gene from the paper and Gen. Bank?
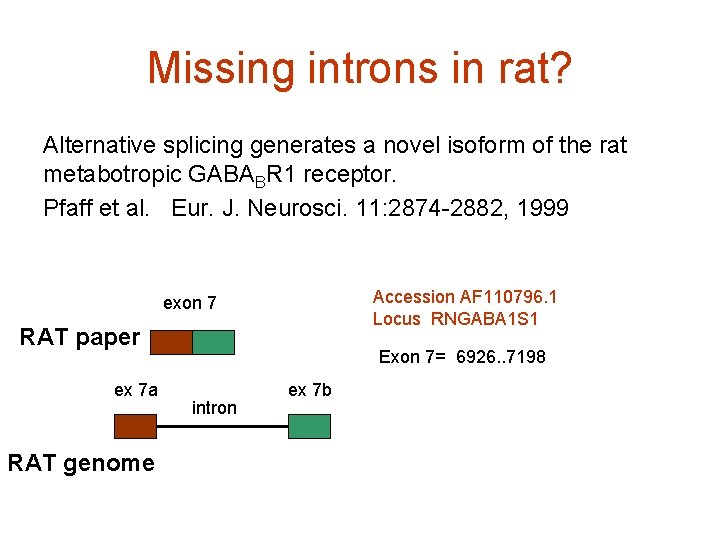
Missing introns in rat? Alternative splicing generates a novel isoform of the rat metabotropic GABABR 1 receptor. Pfaff et al. Eur. J. Neurosci. 11: 2874 -2882, 1999 Accession AF 110796. 1 Locus RNGABA 1 S 1 exon 7 RAT paper ex 7 a RAT genome Exon 7= 6926. . 7198 intron ex 7 b

Error found in study of first ancient African genome http: //www. nature. com/news/error-found-in-study-of-first-ancient-african-genome-1. 19258 This week the authors issued a note explaining the mistake in their October 2015 Science paper on the genome of a 4, 500 year-old man from Ethiopia 1 — the first complete ancient human genome from Africa. The man was named after Mota Cave, where his remains were found. (Incompatible software)
- Slides: 46