APOSYS workshop on data analysis and pathway charting





- Slides: 33

APO-SYS workshop on data analysis and pathway charting Igor Ulitsky Ron Shamir’s Computational Genomics Group

Part I: Presentations Ø EXPANDER Ø AMADEUS Ø SPIKE Ø MATISSE

Part II: Hands-on Session Ø EXPANDER Ø MATISSE Ø SPIKE

EXPression ANalyzer and Display. ER Adi Maron-Katz Chaim Linhart Amos Tanay Rani Elkon Israel Steinfeld Seagull Shavit Igor Ulitsky Roded Sharan Yossi Shiloh Ron Shamir http: //acgt. cs. tau. ac. il/expander

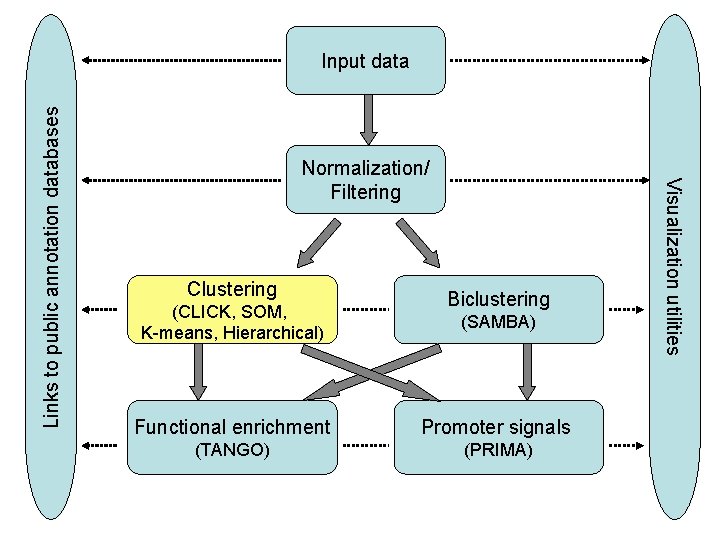
EXPANDER – Low level analysis: • • Missing data estimation (KNN or manual) Normalization: quantile, loess Filtering: fold change, variation, t-test Standardization: mean 0 std 1, take log, fixed norm – High level gene partition analysis: • Clustering • Biclustering – Ascribing biological meaning to patterns: • Enriched functional categories (Gene Ontology) • Identify transcriptional regulators – promoter analysis • Built-in support for 9 organisms: – human, mouse, rat, chicken, zebrafish, fly, worm, arabidopsis, yeast

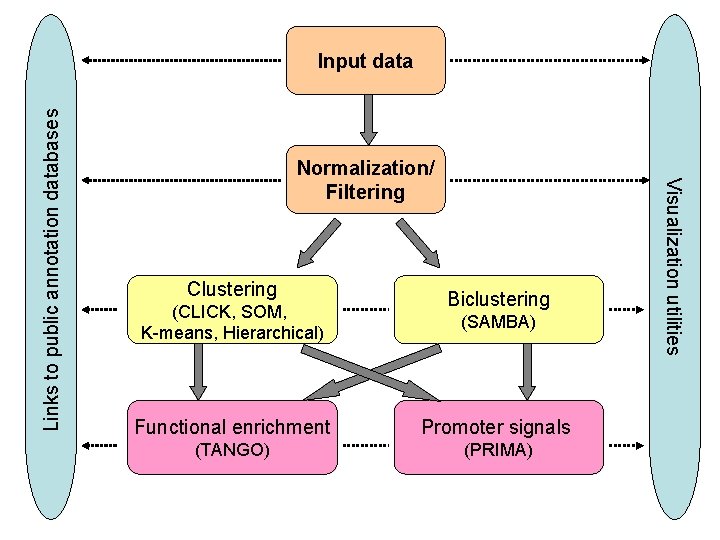
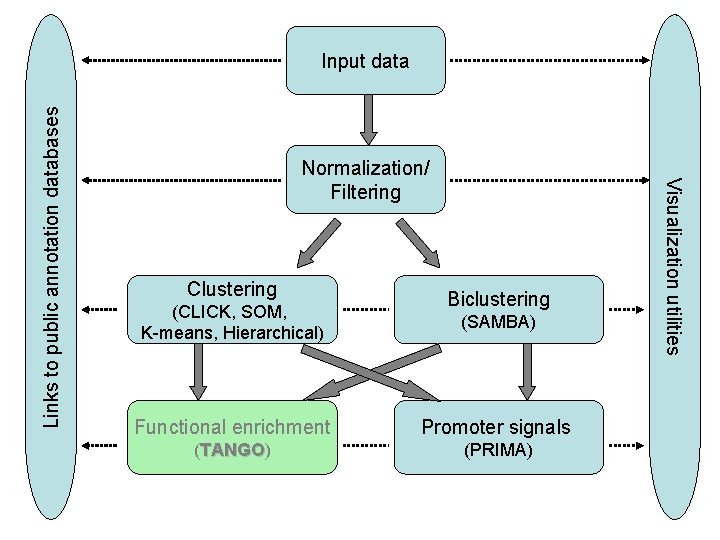
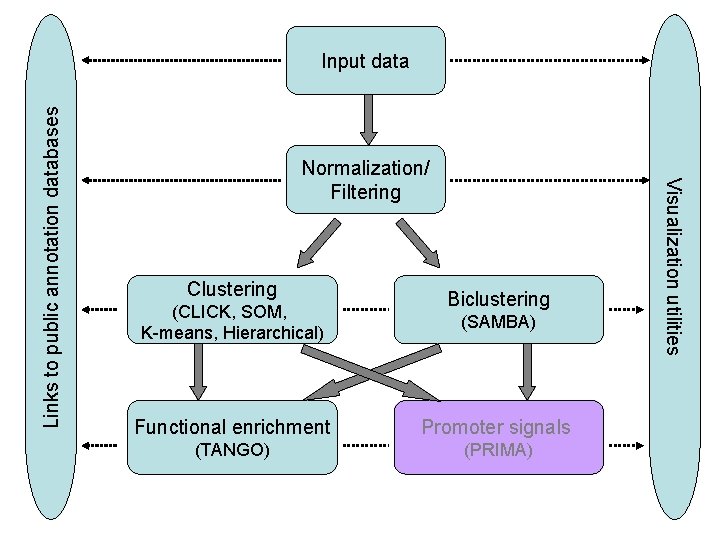
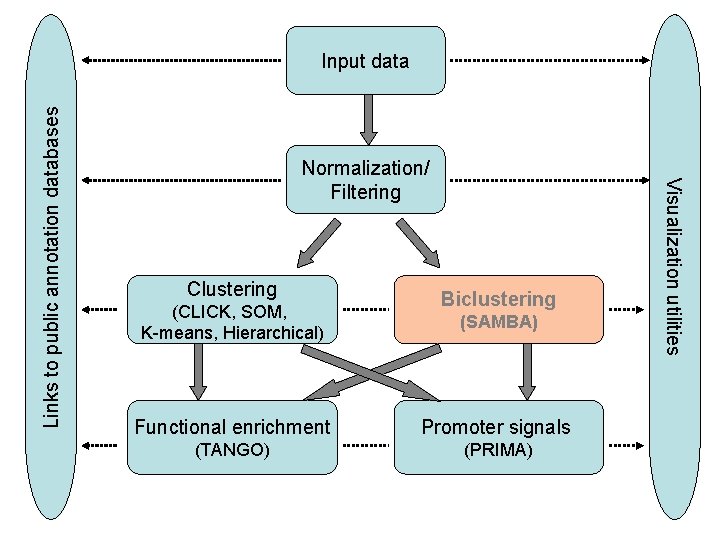
Normalization/ Filtering Clustering (CLICK, SOM, K-means, Hierarchical) Biclustering (SAMBA) Functional enrichment Promoter signals (TANGO) (PRIMA) Visualization utilities Links to public annotation databases Input data

EXPANDER - Preprocessing • Input data: Expression matrix (probe-row; condition-column) • One-channel data (e. g. , Affymetrix) • Dual-channel data (c. DNA microarrays, data are (log) ratios between the Red and Green channels) • ‘. cel’ files ID conversion file: map probes to genes Gene sets data n Data definitions: Defining condition subsets Data type & scale (log)

EXPANDER – Preprocessing (II) § Data Adjustments: Missing value estimation (KNN or arbitrary) Merging conditions Normalization: removal of systematic biases from the analyzed chips § Implemented methods: quantile, lowess § Visualization: box plots, scatter plots (simple, M vs. A)

EXPANDER – Preprocessing (III) § Filtering: Focus downstream analysis on the set of “responding genes” § Fold-Change § Variation § Statistical tests (T-test) § Standardization : Create a common scale § For each probe Mean=0, STD=1 § Log data (base 2) § Fixed Norm (divide by norm of probe vector)

Normalization/ Filtering Clustering (CLICK, SOM, K-means, Hierarchical) Biclustering (SAMBA) Functional enrichment Promoter signals (TANGO) (PRIMA) Visualization utilities Links to public annotation databases Input data

Cluster Analysis • Partition the responding genes into distinct sets, each with a particular expression pattern § Identify major patterns in the data: reduce the dimensionality of the problem § co-expression → co-function § co-expression → co-regulation • Partition the genes to achieve: Ø Homogeneity: genes inside a cluster show highly similar expression pattern. Ø Separation: genes from different clusters have different expression patterns.

Cluster Analysis (II) • Implemented algorithms: – CLICK, K-means, SOM, Hierarchical • Visualization: – – Mean expression patterns Heat-maps

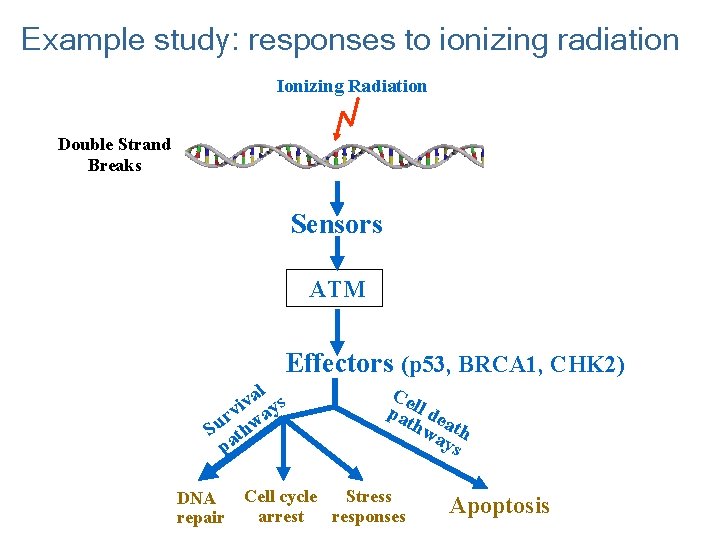
Example study: responses to ionizing radiation Ionizing Radiation Double Strand Breaks Sensors ATM Effectors (p 53, BRCA 1, CHK 2) l a v vi ays r Su thw pa DNA repair Ce pa ll de thw ath ays Cell cycle Stress arrest responses Apoptosis

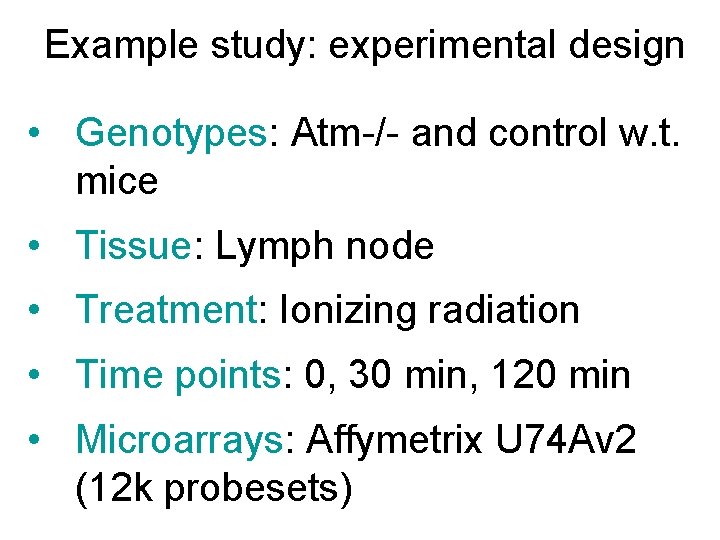
Example study: experimental design • Genotypes: Atm-/- and control w. t. mice • Tissue: Lymph node • Treatment: Ionizing radiation • Time points: 0, 30 min, 120 min • Microarrays: Affymetrix U 74 Av 2 (12 k probesets)

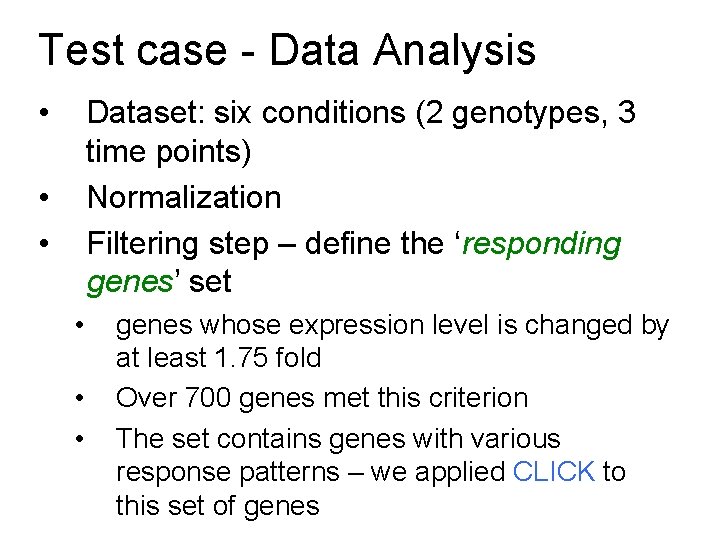
Test case - Data Analysis • Dataset: six conditions (2 genotypes, 3 time points) Normalization Filtering step – define the ‘responding genes’ set • • • genes whose expression level is changed by at least 1. 75 fold Over 700 genes met this criterion The set contains genes with various response patterns – we applied CLICK to this set of genes

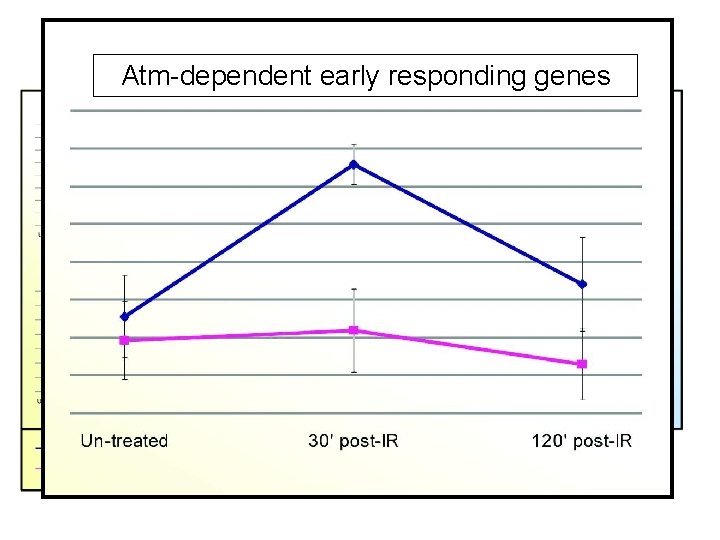
Major. Atm-dependent Gene Clusters –early Irradiated Lymph node responding genes

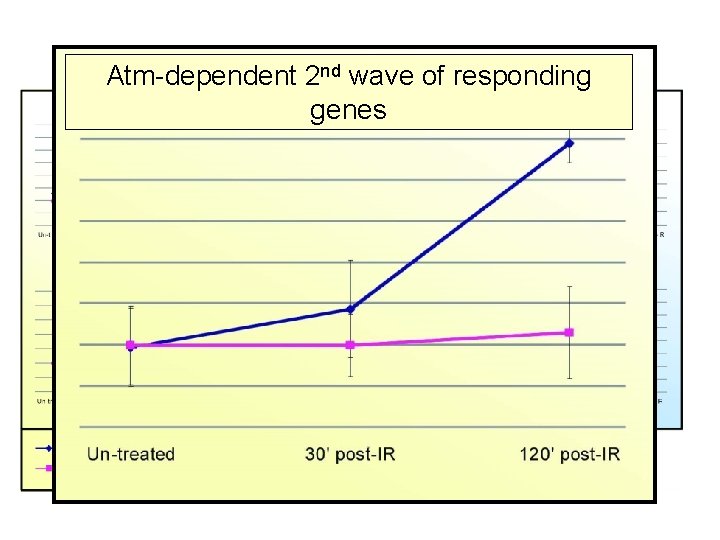
Major Gene Clusters 2–nd. Irradiated Lymph node Atm-dependent wave of responding genes

Normalization/ Filtering Clustering (CLICK, SOM, K-means, Hierarchical) Biclustering (SAMBA) Functional enrichment Promoter signals (TANGO) TANGO (PRIMA) Visualization utilities Links to public annotation databases Input data

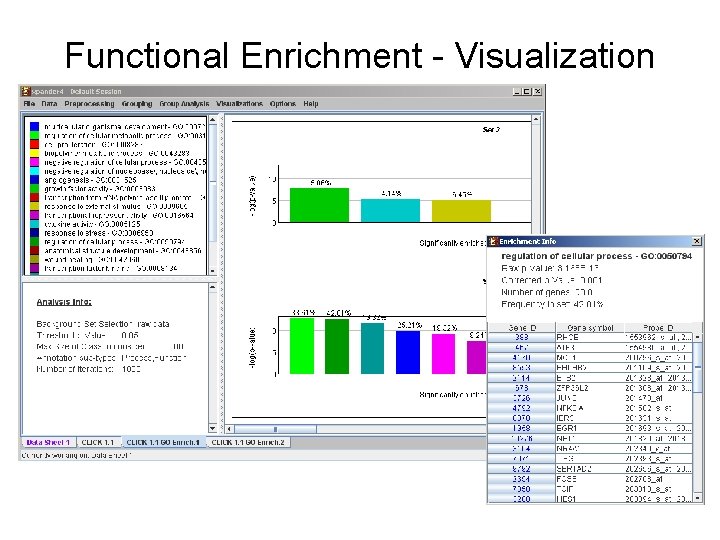
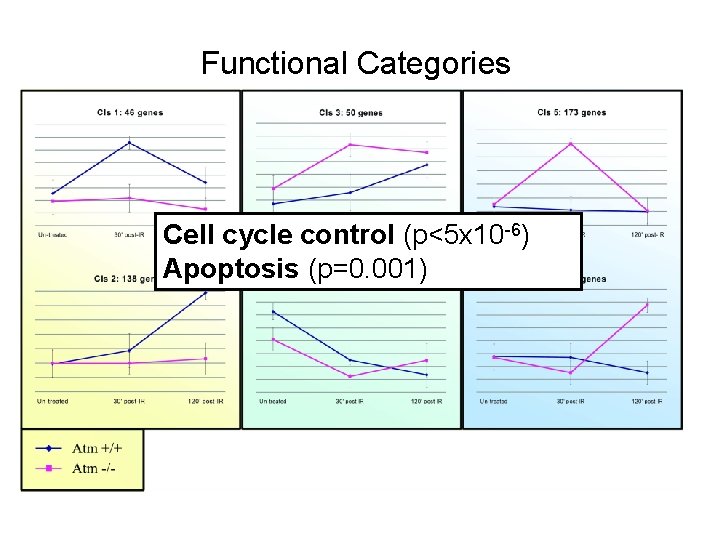
Ascribe Functional Meaning to the Clusters • Gene Ontology (GO) annotations for human, mouse, rat, chicken, fly, worm, Arabidopsis, Zebrafish and yeast. • TANGO: Apply statistical tests that seek over-represented GO functional categories in the clusters.

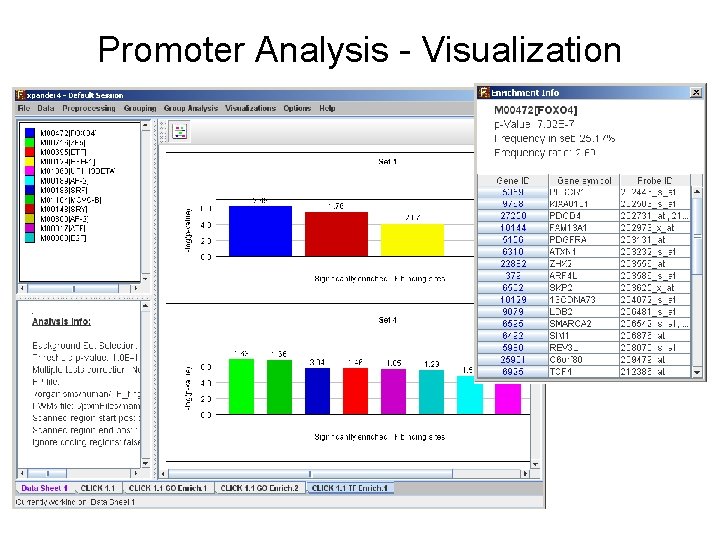
Functional Enrichment - Visualization

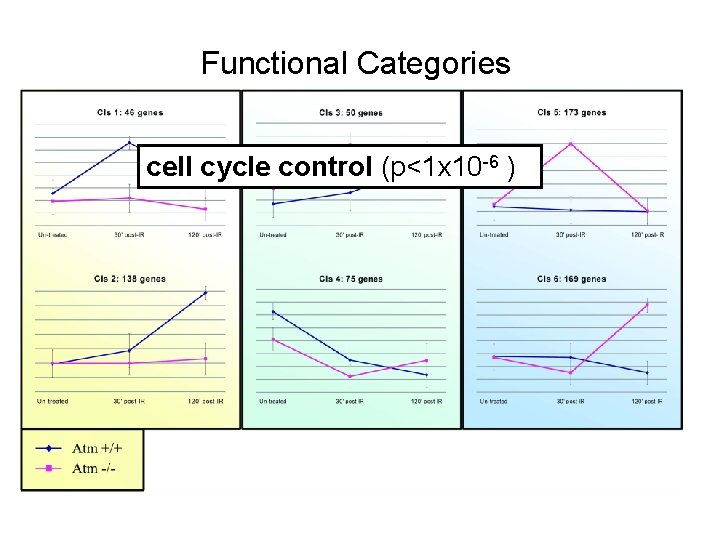
Functional Categories cell cycle control (p<1 x 10 -6 )

Functional Categories Cell cycle control (p<5 x 10 -6) Apoptosis (p=0. 001)

Normalization/ Filtering Clustering (CLICK, SOM, K-means, Hierarchical) Biclustering (SAMBA) Functional enrichment Promoter signals (TANGO) (PRIMA) Visualization utilities Links to public annotation databases Input data

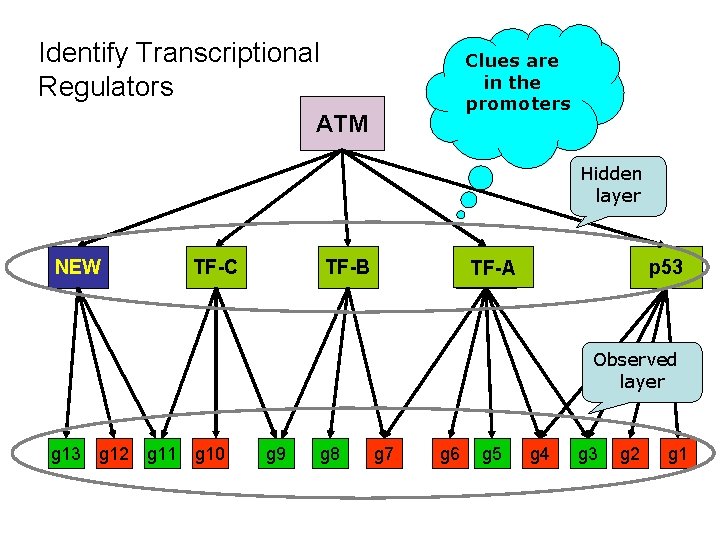
Identify Transcriptional Regulators Clues are in the promoters ATM Hidden layer NEW ? TF-C ? TF-B ? ? TF-A p 53 ? Observed layer g 13 g 12 g 11 g 10 g 9 g 8 g 7 g 6 g 5 g 4 g 3 g 2 g 1

‘Reverse engineering’ of transcriptional networks • Infers regulatory mechanisms from gene expression data – Assumption: co-expression → transcriptional co-regulation → common cis-regulatory promoter elements • Step 1: Identification of co-expressed genes using microarray technology (clustering algs) • Step 2: Computational identification of cisregulatory elements that are over-represented in promoters of the co-expressed gene

PRIMA – general description • Input: – Target set (e. g. , co-expressed genes) – Background set (e. g. , all genes on the chip) • Analysis: – Identify transcription factors whose binding site signatures are enriched in the ‘Target set’ with respect to the ‘Background set’. • TF binding site models – TRANSFAC DB • Default: From -1000 bp to 200 bp relative the TSS

Promoter Analysis - Visualization

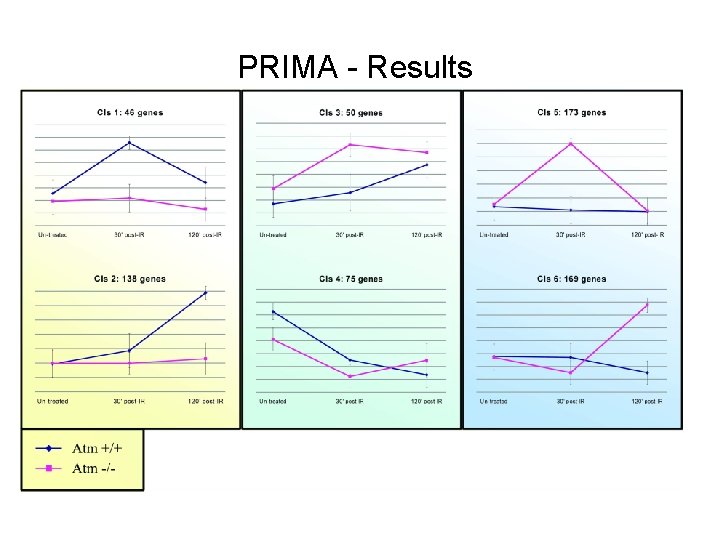
PRIMA - Results

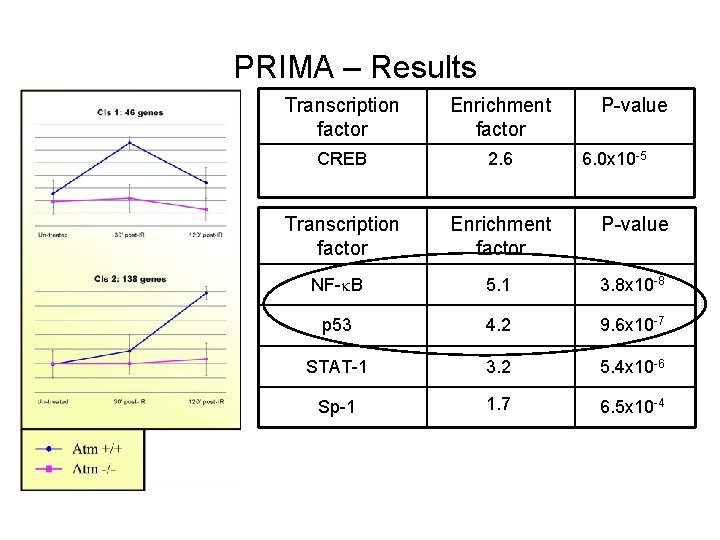
PRIMA – Results Transcription factor Enrichment factor P-value CREB 2. 6 Transcription factor Enrichment factor P-value NF- B 5. 1 3. 8 x 10 -8 p 53 4. 2 9. 6 x 10 -7 STAT-1 3. 2 5. 4 x 10 -6 Sp-1 1. 7 6. 5 x 10 -4 6. 0 x 10 -5

Normalization/ Filtering Clustering (CLICK, SOM, K-means, Hierarchical) Biclustering (SAMBA) Functional enrichment Promoter signals (TANGO) (PRIMA) Visualization utilities Links to public annotation databases Input data

Biclustering § Clustering becomes too restrictive on large datasets: • Seeks global partition of genes according to similarity in their expression across ALL conditions § Relevant knowledge can be revealed by identifying genes with common pattern across a subset of the conditions • Biclustering algorithmic approach

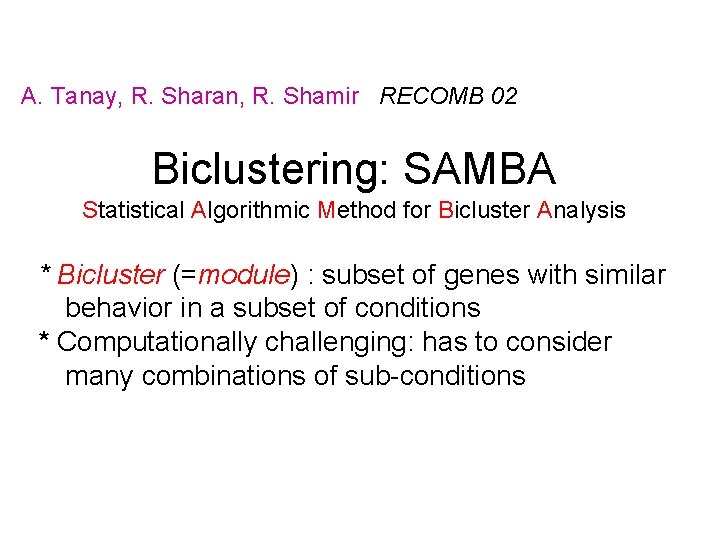
A. Tanay, R. Sharan, R. Shamir RECOMB 02 Biclustering: SAMBA Statistical Algorithmic Method for Bicluster Analysis * Bicluster (=module) : subset of genes with similar behavior in a subset of conditions * Computationally challenging: has to consider many combinations of sub-conditions

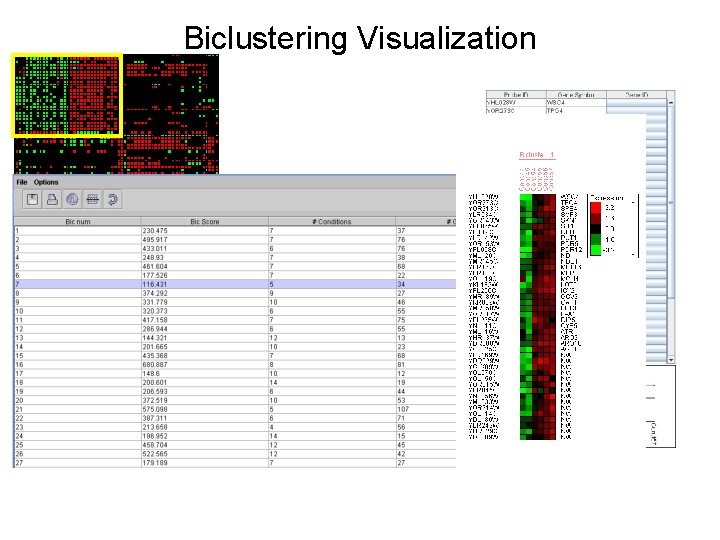
Biclustering Visualization