Any Kd you like outline Two ITC experiments









- Slides: 27

Any Kd you like – outline • Two ITC experiments • The problem for Biophysics • Problem data - literature and in-house examples • Interpreting biphasic data © Astex Pharmaceuticals, Inc

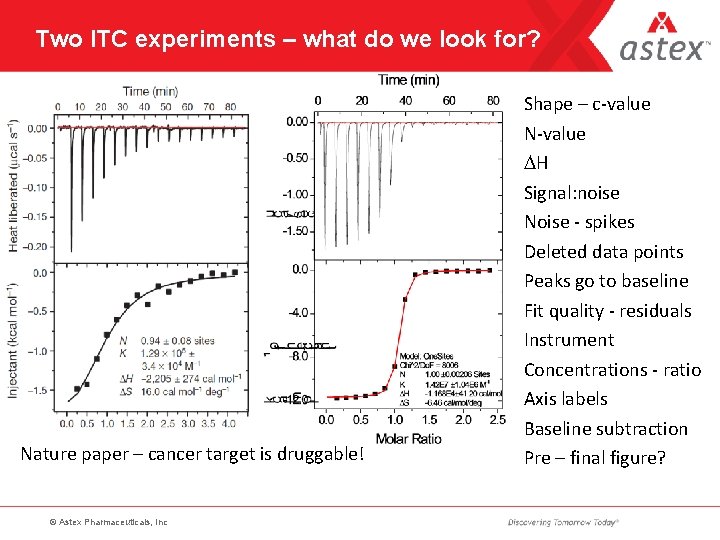
Two ITC experiments – what do we look for? Nature paper – cancer target is druggable! © Astex Pharmaceuticals, Inc Shape – c-value N-value DH Signal: noise Noise - spikes Deleted data points Peaks go to baseline Fit quality - residuals Instrument Concentrations - ratio Axis labels Baseline subtraction Pre – final figure?

Science literature reproducibility • Amgen – reproducibility of “landmark” articles in cancer research – papers describing something new, top journals, reputable labs – 11% reproducible (6/53) • Bayer – 67 validation projects based on exciting published data – in almost two-thirds of the projects, there were inconsistencies between published data and in-house data • Details of studies not published but alarming numbers – – inappropriate use of statistics limits of preclinical models selective use of data poor study design • Strange bedfellows – cell biology and biophysics © Astex Pharmaceuticals, Inc

The problem for Biophysics • A fictitious example: • New cancer target/pathway identified and shown to be druggable – virtual screen against target X – some compounds show activity in alpha screen assay – one particular compound (Y ) is active in cells and pulls down target X – confirm that compound Y interacts with protein X using SPR/NMR/Tm shift/MST – usually these sort of papers have no x-ray crystal structures or ITC • The problem for Biophysics – why can’t you see it binding? © Astex Pharmaceuticals, Inc

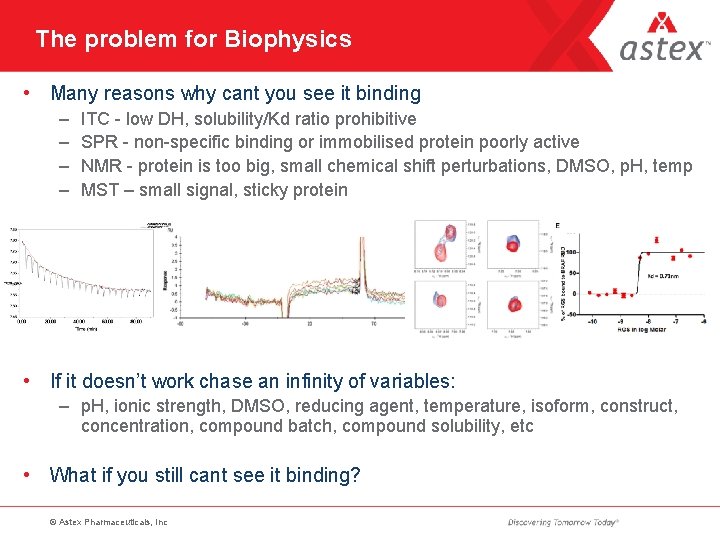
The problem for Biophysics • Many reasons why cant you see it binding – – ITC - low DH, solubility/Kd ratio prohibitive SPR - non-specific binding or immobilised protein poorly active NMR - protein is too big, small chemical shift perturbations, DMSO, p. H, temp MST – small signal, sticky protein • If it doesn’t work chase an infinity of variables: – p. H, ionic strength, DMSO, reducing agent, temperature, isoform, construct, concentration, compound batch, compound solubility, etc • What if you still cant see it binding? © Astex Pharmaceuticals, Inc

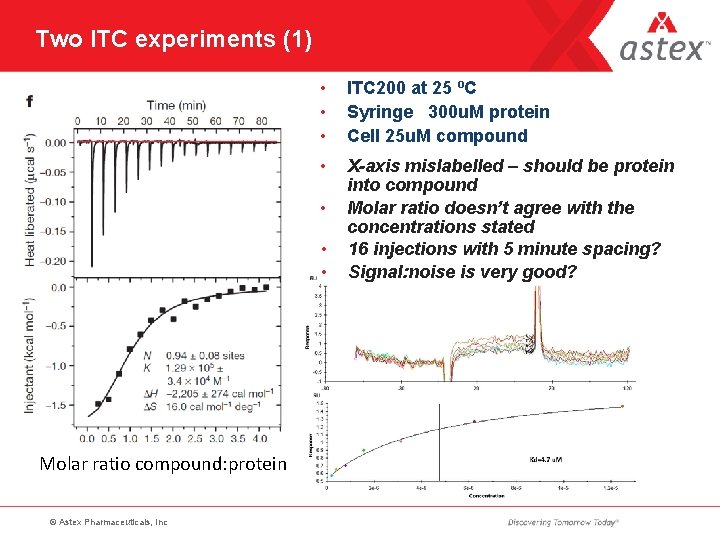
Two ITC experiments (1) • • • ITC 200 at 25 ºC Syringe 300 u. M protein Cell 25 u. M compound • X-axis mislabelled – should be protein into compound Molar ratio doesn’t agree with the concentrations stated 16 injections with 5 minute spacing? Signal: noise is very good? • • • Molar ratio compound: protein © Astex Pharmaceuticals, Inc

ITC – the gold standard • Most published ITC data is good quality – – fit goes through data stoichiometry makes sense units/experimental parameters fine (we all make mistakes) • Usually obvious when ITC data is poor, or fitting is hallucinogenic – why it gets through the referees is another matter • It can be difficult to interpret some titrations – low DH, solubility: Kd (can’t saturate), drifting baseline, inactive protein • Examples in-house and literature: © Astex Pharmaceuticals, Inc

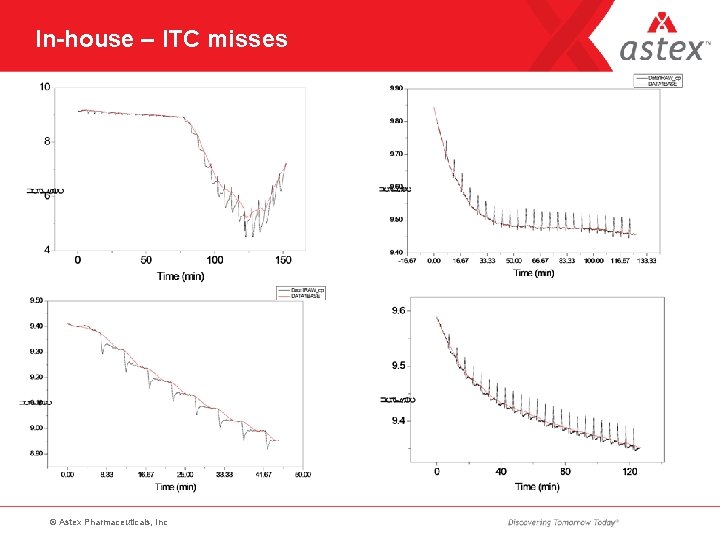
In-house – ITC misses © Astex Pharmaceuticals, Inc

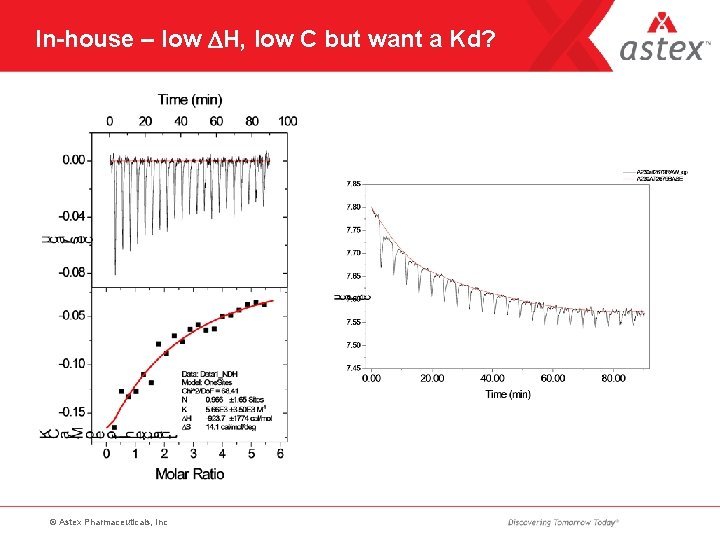
In-house – low DH, low C but want a Kd? © Astex Pharmaceuticals, Inc

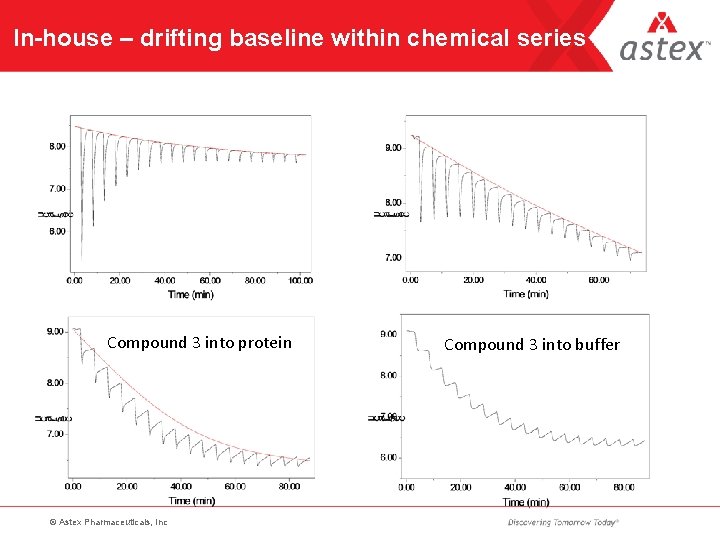
In-house – drifting baseline within chemical series Compound 3 into protein © Astex Pharmaceuticals, Inc Compound 3 into buffer

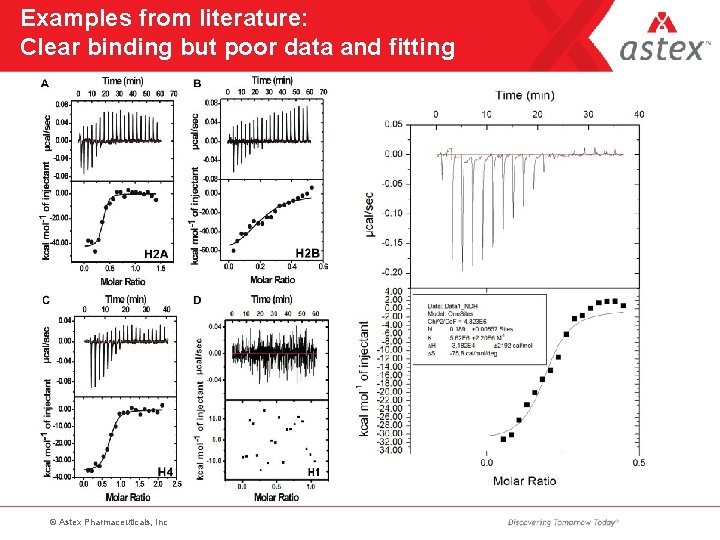
Examples from literature: Clear binding but poor data and fitting © Astex Pharmaceuticals, Inc

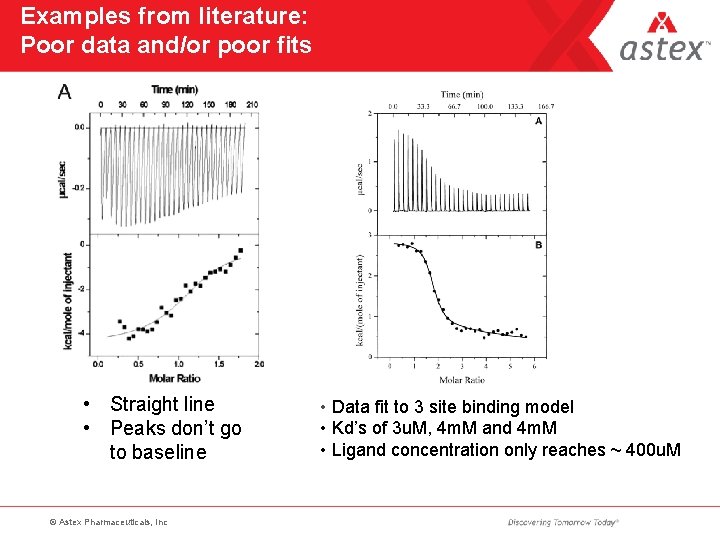
Examples from literature: Poor data and/or poor fits • Straight line • Peaks don’t go to baseline © Astex Pharmaceuticals, Inc • Data fit to 3 site binding model • Kd’s of 3 u. M, 4 m. M and 4 m. M • Ligand concentration only reaches ~ 400 u. M

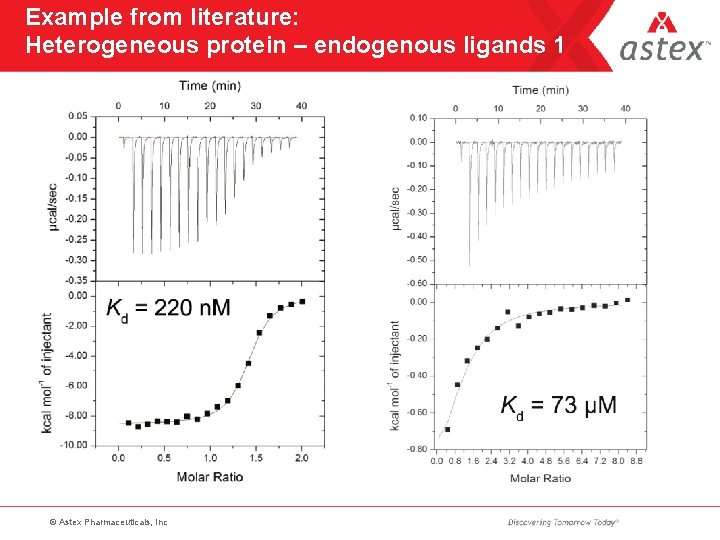
Example from literature: Heterogeneous protein – endogenous ligands 1 © Astex Pharmaceuticals, Inc

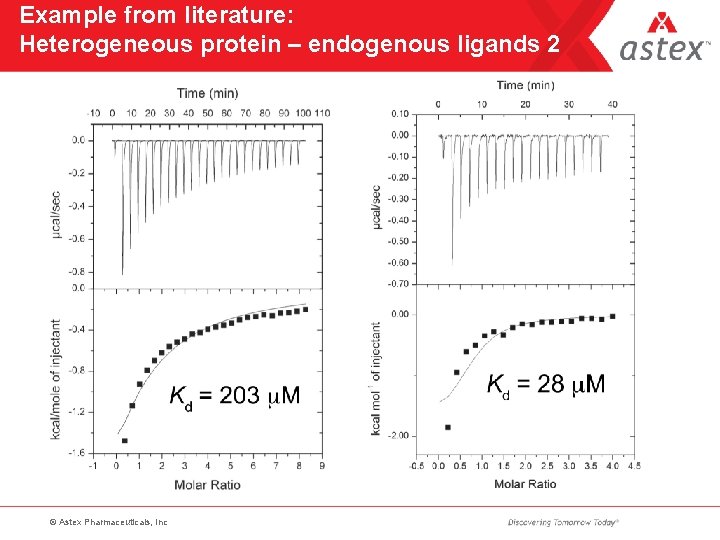
Example from literature: Heterogeneous protein – endogenous ligands 2 © Astex Pharmaceuticals, Inc

Endogenous ligands • Remarkable how many proteins copurify with bound material – peptides – co-factors – previously unknown ligands • An accurate N-value is an invaluable protein QC parameter • Native and LCMS can help identify endogenous material • Helps explain poor ITC fits © Astex Pharmaceuticals, Inc

Two ITC experiments – (2) © Astex Pharmaceuticals, Inc

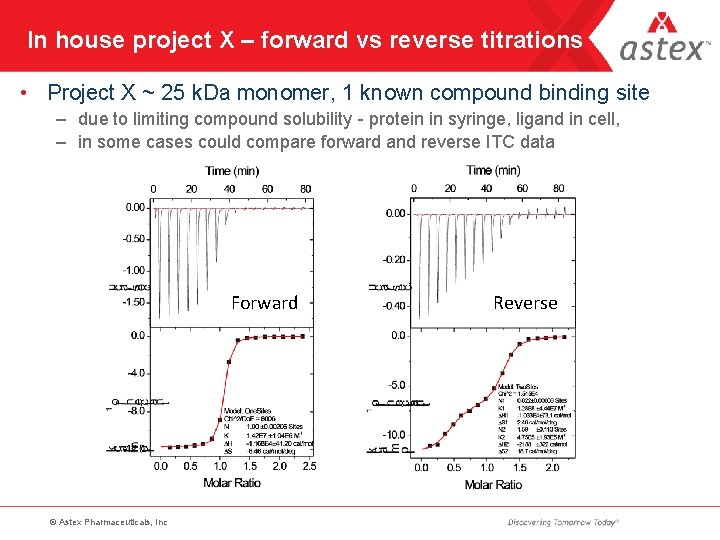
In house project X – forward vs reverse titrations • Project X ~ 25 k. Da monomer, 1 known compound binding site – due to limiting compound solubility - protein in syringe, ligand in cell, – in some cases could compare forward and reverse ITC data Forward © Astex Pharmaceuticals, Inc Reverse

Biphasic ITC data – previous in-house examples Mixture of enantiomers © Astex Pharmaceuticals, Inc Oligomer – positive cooperativity 2 protein states? 2 binding sites? Compound mixture?

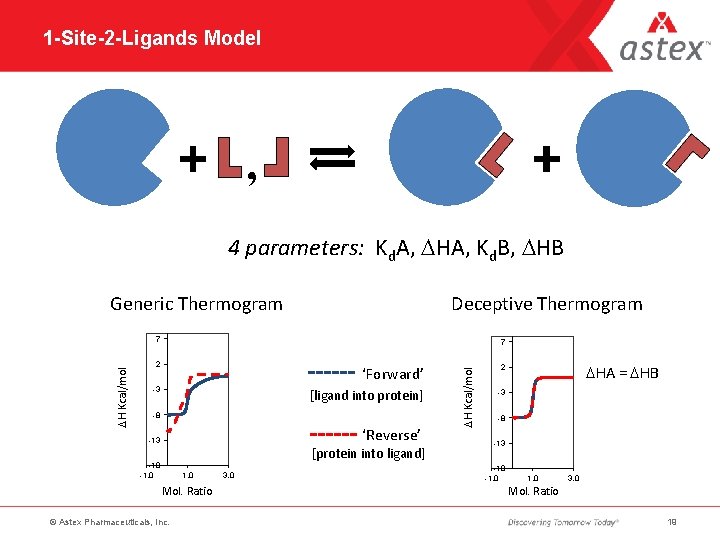
1 -Site-2 -Ligands Model , 4 parameters: Kd. A, DHA, Kd. B, DHB Generic Thermogram Deceptive Thermogram 7 2 ‘Forward’ -3 [ligand into protein] -8 ‘Reverse’ -13 [protein into ligand] -18 -1. 0 Mol. Ratio © Astex Pharmaceuticals, Inc. 3. 0 DH Kcal/mol 7 DHA = DHB 2 -3 -8 -13 -18 -1. 0 3. 0 Mol. Ratio 19

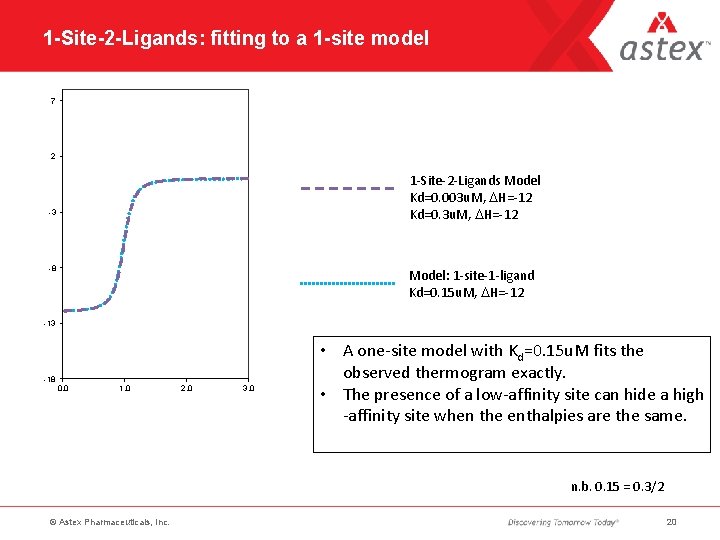
1 -Site-2 -Ligands: fitting to a 1 -site model 7 2 1 -Site-2 -Ligands Model Kd=0. 003 u. M, DH=-12 Kd=0. 3 u. M, DH=-12 -3 -8 Model: 1 -site-1 -ligand Kd=0. 15 u. M, DH=-12 -13 -18 0. 0 1. 0 2. 0 3. 0 • A one-site model with Kd=0. 15 u. M fits the observed thermogram exactly. • The presence of a low-affinity site can hide a high -affinity site when the enthalpies are the same. n. b. 0. 15 = 0. 3/2 © Astex Pharmaceuticals, Inc. 20

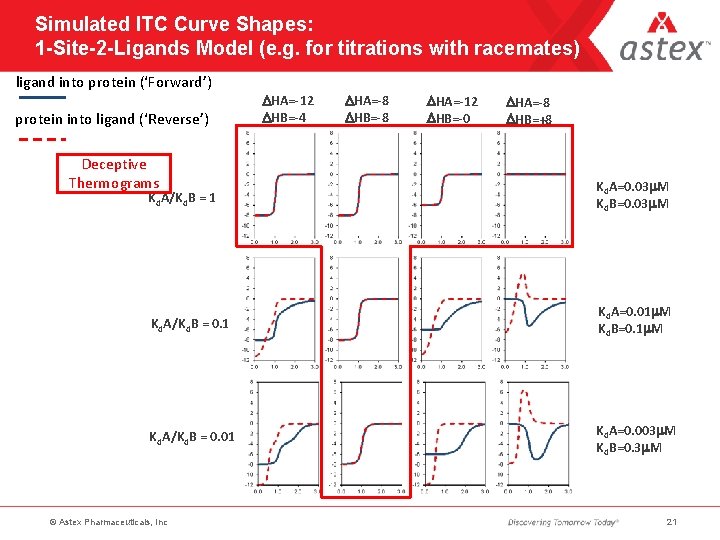
Simulated ITC Curve Shapes: 1 -Site-2 -Ligands Model (e. g. for titrations with racemates) ligand into protein (‘Forward’) protein into ligand (‘Reverse’) Deceptive Thermograms DHA=-12 DHB=-4 DHA=-8 DHB=-8 DHA=-12 DHB=-0 DHA=-8 DHB=+8 Kd. A/Kd. B = 1 Kd. A=0. 03 m. M Kd. B=0. 03 m. M Kd. A/Kd. B = 0. 1 Kd. A=0. 01 m. M Kd. B=0. 1 m. M Kd. A/Kd. B = 0. 01 Kd. A=0. 003 m. M Kd. B=0. 3 m. M © Astex Pharmaceuticals, Inc 21

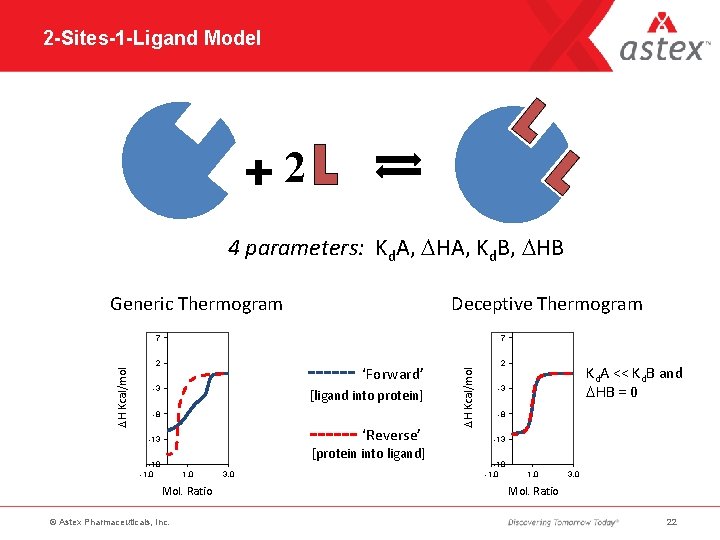
2 -Sites-1 -Ligand Model 2 4 parameters: Kd. A, DHA, Kd. B, DHB Generic Thermogram Deceptive Thermogram 7 2 ‘Forward’ -3 [ligand into protein] -8 ‘Reverse’ -13 [protein into ligand] -18 -1. 0 Mol. Ratio © Astex Pharmaceuticals, Inc. 3. 0 DH Kcal/mol 7 2 Kd. A << Kd. B and DHB = 0 -3 -8 -13 -18 -1. 0 3. 0 Mol. Ratio 22

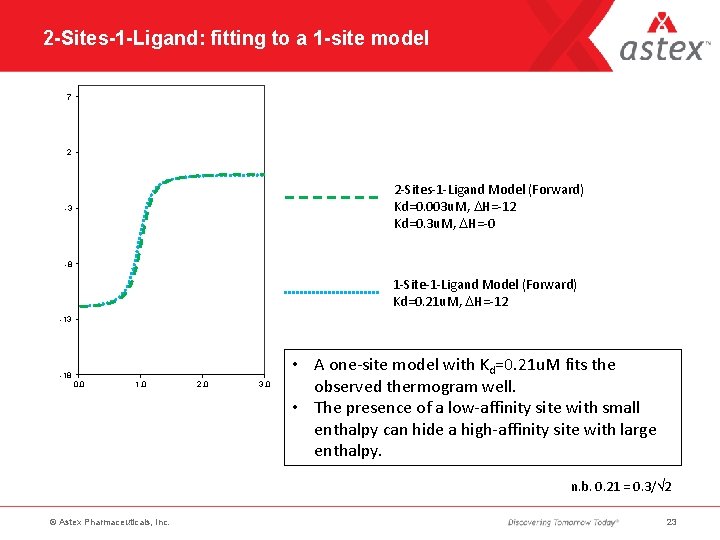
2 -Sites-1 -Ligand: fitting to a 1 -site model 7 2 2 -Sites-1 -Ligand Model (Forward) Kd=0. 003 u. M, DH=-12 Kd=0. 3 u. M, DH=-0 -3 -8 1 -Site-1 -Ligand Model (Forward) Kd=0. 21 u. M, DH=-12 -13 -18 0. 0 1. 0 2. 0 3. 0 • A one-site model with Kd=0. 21 u. M fits the observed thermogram well. • The presence of a low-affinity site with small enthalpy can hide a high-affinity site with large enthalpy. n. b. 0. 21 = 0. 3/ 2 © Astex Pharmaceuticals, Inc. 23

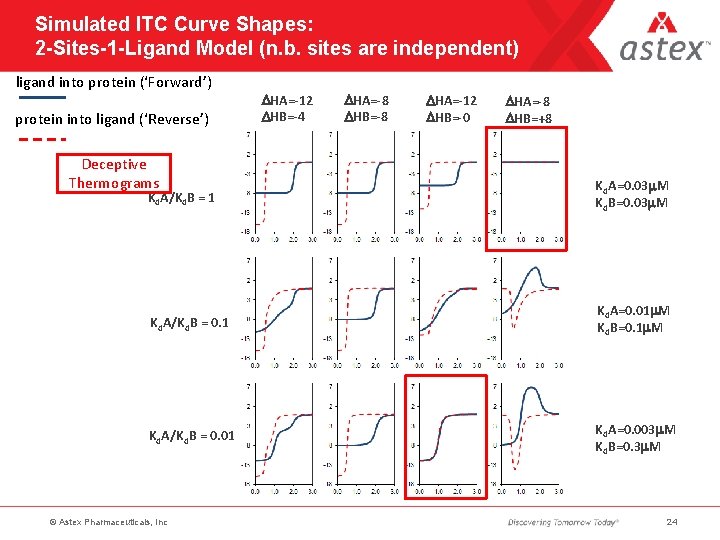
Simulated ITC Curve Shapes: 2 -Sites-1 -Ligand Model (n. b. sites are independent) ligand into protein (‘Forward’) protein into ligand (‘Reverse’) Deceptive Thermograms DHA=-12 DHB=-4 DHA=-8 DHB=-8 DHA=-12 DHB=-0 DHA=-8 DHB=+8 Kd. A/Kd. B = 1 Kd. A=0. 03 m. M Kd. B=0. 03 m. M Kd. A/Kd. B = 0. 1 Kd. A=0. 01 m. M Kd. B=0. 1 m. M Kd. A/Kd. B = 0. 01 Kd. A=0. 003 m. M Kd. B=0. 3 m. M © Astex Pharmaceuticals, Inc 24

What do we believe? • We do not yet have an appropriate model to fit – fitting to a 2 -site model does not give consistent values for Kd 1, Kd 2, DH 1, DH 2 or the stoichiometry of the interaction, for titrations using different methods or under different conditions – Additional parameters or equilibria may be required (e. g. cooperativity between the sites or protein oligomerisation) • If the final model includes two ligand binding sites they are likely to have very different affinities (~10 n. M/ ~1 u. M) • If two sites are involved, the weaker site may be for the observed cellular activity

ITC – still remains the gold standard • ITC is the most plausible published affinity measurement – but doesn’t mean that its infallible • If data doesn’t fit well to 1: 1 model it is telling you something – – – although it isn't always easy to figure out what reverse titrations can help confirm particular models don’t blindly throw a 1: 1 fit at all data N-value has to be understood raw data should be shown? • More software options – better baseline algorithms – danger of data processing becoming a black-box – important to get hands on © Astex Pharmaceuticals, Inc

Acknowledgements • Amanda Price • Sue Saalau • Glyn Williams © Astex Pharmaceuticals, Inc