Antioxidant Enzymes Maria Holmstrom Qiang Zhang Nicole Milkovic







- Slides: 23

Antioxidant Enzymes Maria Holmstrom Qiang Zhang Nicole Milkovic Erin Rosenbaugh

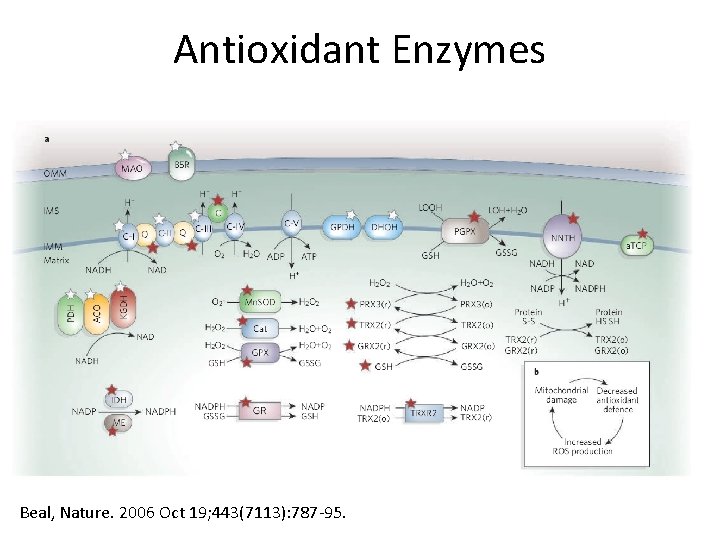
Introduction to Antioxidant Enzymes Beal, Nature. 2006 Oct 19; 443(7113): 787 -95.

Superoxide dismutases • Catalyzes the dismutation of superoxide into oxygen and hydrogen peroxide – Diffusion limited • Found in nearly all oxygen-exposed cells • Categorized by metal prosthetic group – Cu/Zn, Mn, Fe or Ni

Species • B. Subtilis • E. Coli Mn. SOD (sod. A) and Fe. SOD (sod. B) • S. Cerevisiae Cu. Zn. SOD (sod 1) and Mn. SOD (sod 2) • H. Sapiens Cu. Zn. SOD (sod 1), Mn. SOD (sod 2) and EC-SOD (Cu. Zn, sod 3)

Localization • Bacteria – SOD A cytoplasm – SOD B cytoplasm • Eukaryotes – SOD 1 cytosol – SOD 2 mitochondrial matrix – SOD 3 (humans) glycated and secreted into the extracellular space, and subsequently anchored to heparan sulfate proteoglycans

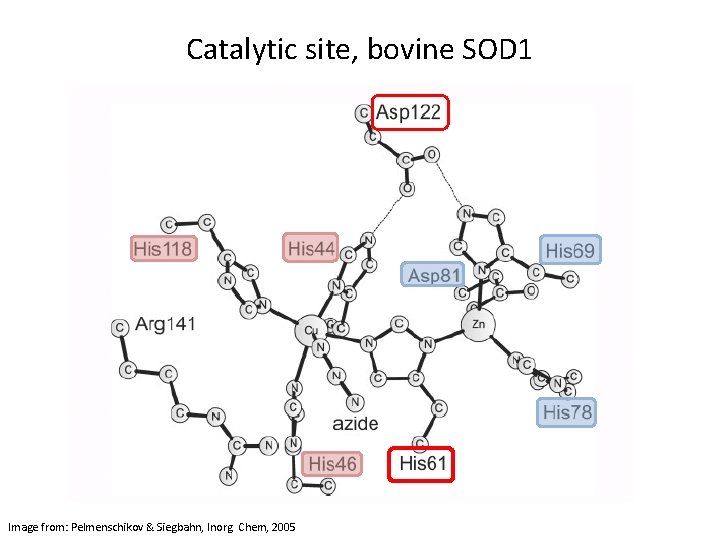
Catalytic site, bovine SOD 1 Image from: Pelmenschikov & Siegbahn, Inorg. Chem, 2005

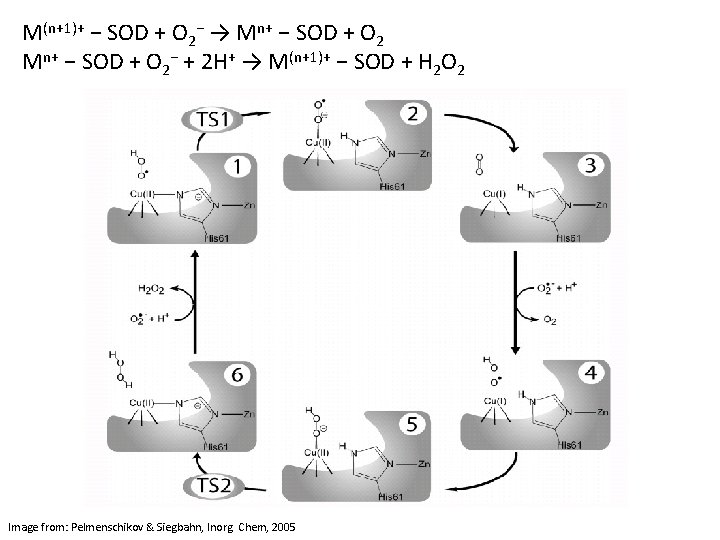
M(n+1)+ − SOD + O 2− → Mn+ − SOD + O 2− + 2 H+ → M(n+1)+ − SOD + H 2 O 2 Image from: Pelmenschikov & Siegbahn, Inorg. Chem, 2005

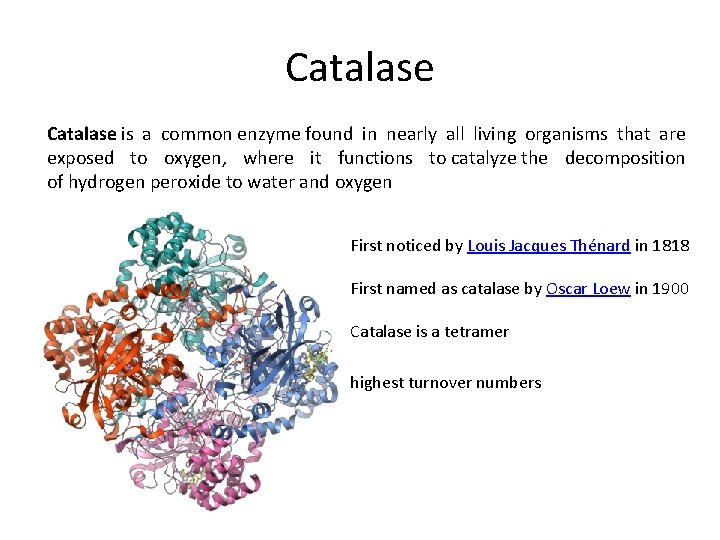
Catalase is a common enzyme found in nearly all living organisms that are exposed to oxygen, where it functions to catalyze the decomposition of hydrogen peroxide to water and oxygen First noticed by Louis Jacques Thénard in 1818 First named as catalase by Oscar Loew in 1900 Catalase is a tetramer highest turnover numbers

Cofactors • Heme • Manganese

Distribution among organisms • All known animals use catalase in every organ, with particularly high concentrations occurring in the liver • Catalase is also universal among plants, and many fungi are also high producers of the enzyme • Catalase has also been observed in some anaerobic microorganisms

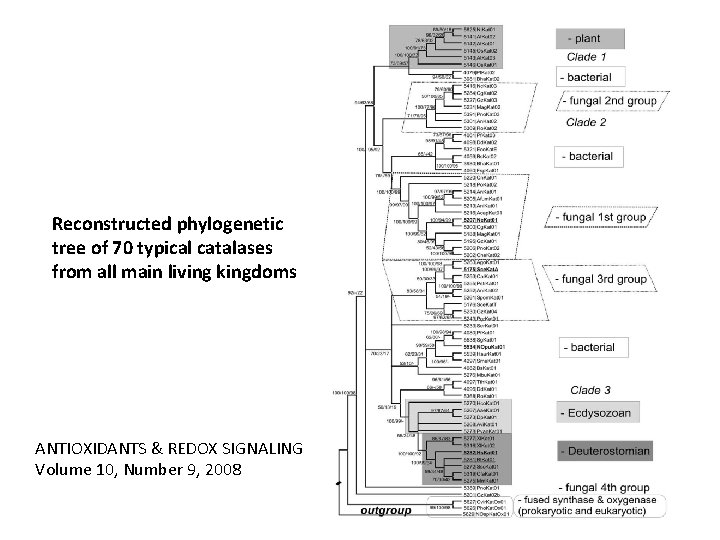
Reconstructed phylogenetic tree of 70 typical catalases from all main living kingdoms ANTIOXIDANTS & REDOX SIGNALING Volume 10, Number 9, 2008

Catalase genes • Bacillus subtilis: kat. A(vegetative catalase 1), kat. X(catalase in spores), kat. E(catalase 2) • E. Coli: kat. E(HPII(III)), kat. G(HPI), kat. P(EHECcatalase) • S. Cerevisiae: CTA 1(Catalase A), CTT 1(Ctt 1 p ) • H. Sapiens: CAT(Catalase)

Location • • Intracellular Extracellular Cell surface Periplasm Cytosol Glyoxysome Mitochondrion

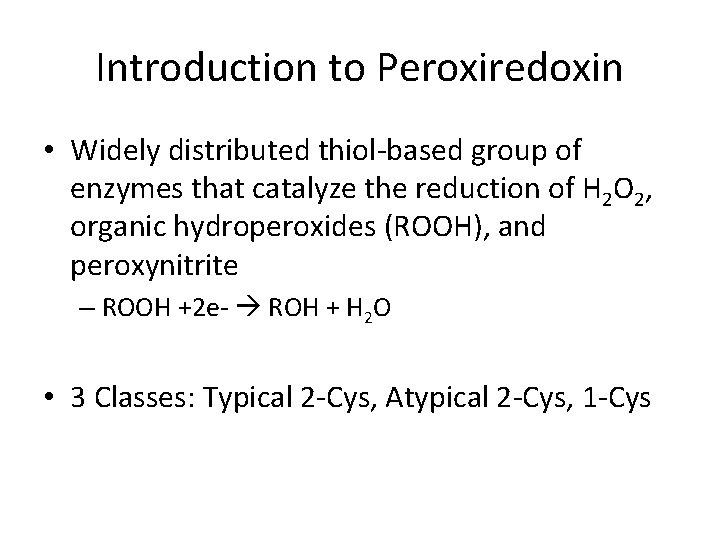
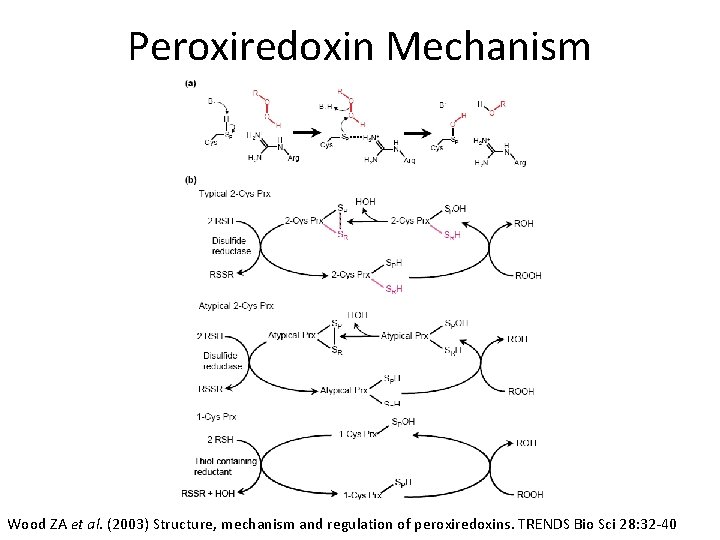
Introduction to Peroxiredoxin • Widely distributed thiol-based group of enzymes that catalyze the reduction of H 2 O 2, organic hydroperoxides (ROOH), and peroxynitrite – ROOH +2 e- ROH + H 2 O • 3 Classes: Typical 2 -Cys, Atypical 2 -Cys, 1 -Cys

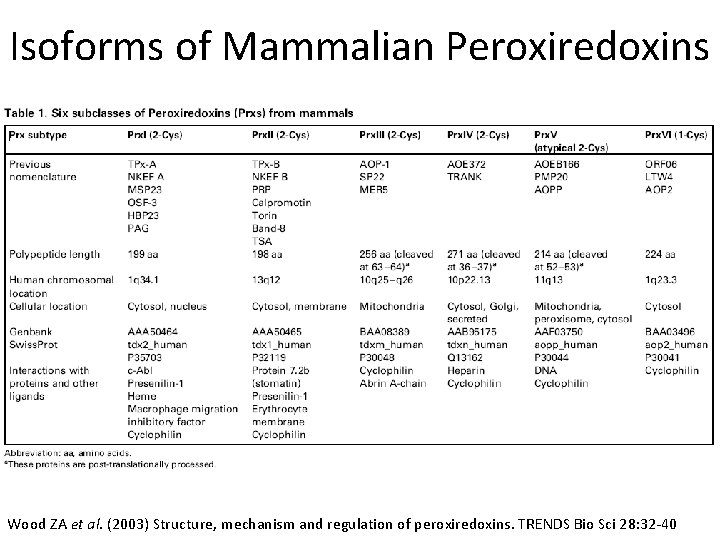
Isoforms of Mammalian Peroxiredoxins Wood ZA et al. (2003) Structure, mechanism and regulation of peroxiredoxins. TRENDS Bio Sci 28: 32 -40

Peroxiredoxin Mechanism Wood ZA et al. (2003) Structure, mechanism and regulation of peroxiredoxins. TRENDS Bio Sci 28: 32 -40

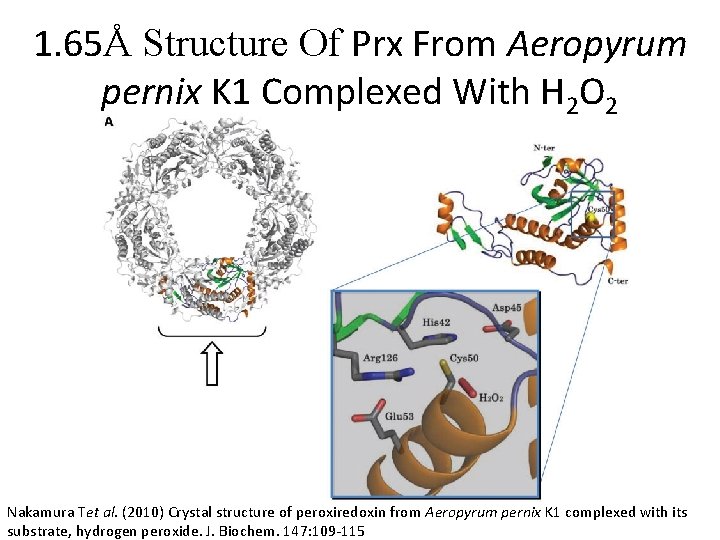
1. 65Å Structure Of Prx From Aeropyrum pernix K 1 Complexed With H 2 O 2 Nakamura Tet al. (2010) Crystal structure of peroxiredoxin from Aeropyrum pernix K 1 complexed with its substrate, hydrogen peroxide. J. Biochem. 147: 109 -115

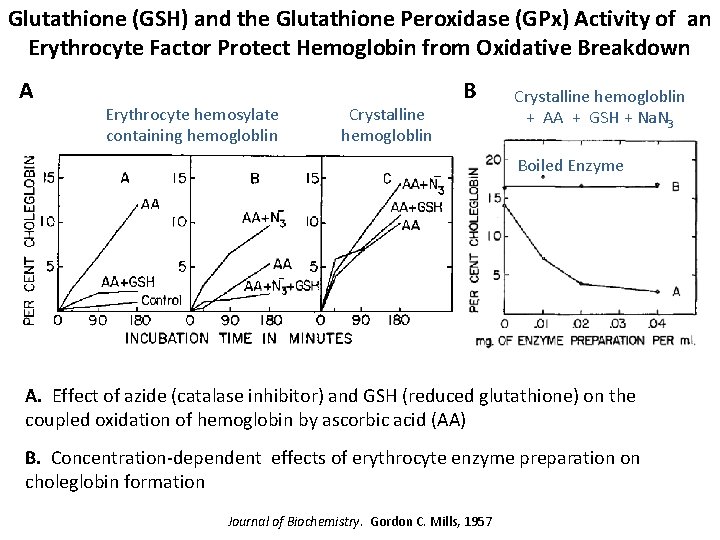
Glutathione (GSH) and the Glutathione Peroxidase (GPx) Activity of an Erythrocyte Factor Protect Hemoglobin from Oxidative Breakdown A Erythrocyte hemosylate containing hemogloblin Crystalline hemogloblin B Crystalline hemogloblin + AA + GSH + Na. N 3 Boiled Enzyme A. Effect of azide (catalase inhibitor) and GSH (reduced glutathione) on the coupled oxidation of hemoglobin by ascorbic acid (AA) B. Concentration-dependent effects of erythrocyte enzyme preparation on choleglobin formation Journal of Biochemistry. Gordon C. Mills, 1957

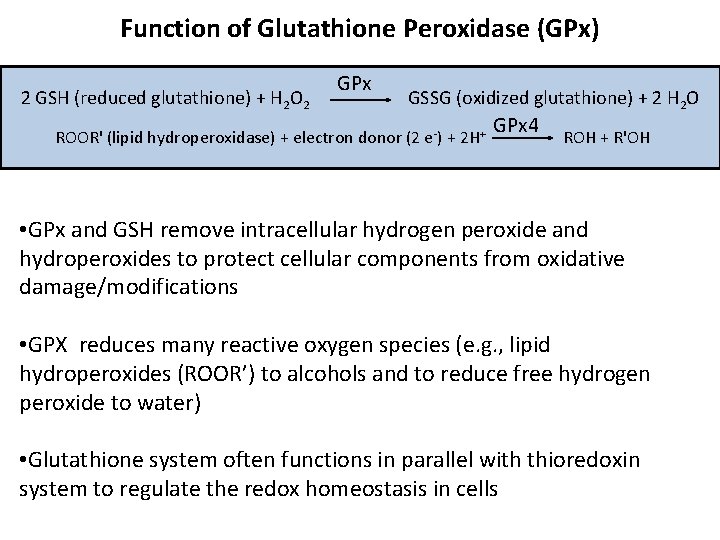
Function of Glutathione Peroxidase (GPx) 2 GSH (reduced glutathione) + H 2 O 2 GPx GSSG (oxidized glutathione) + 2 H 2 O ROOR' (lipid hydroperoxidase) + electron donor (2 e-) + 2 H+ GPx 4 ROH + R'OH • GPx and GSH remove intracellular hydrogen peroxide and hydroperoxides to protect cellular components from oxidative damage/modifications • GPX reduces many reactive oxygen species (e. g. , lipid hydroperoxides (ROOR’) to alcohols and to reduce free hydrogen peroxide to water) • Glutathione system often functions in parallel with thioredoxin system to regulate the redox homeostasis in cells

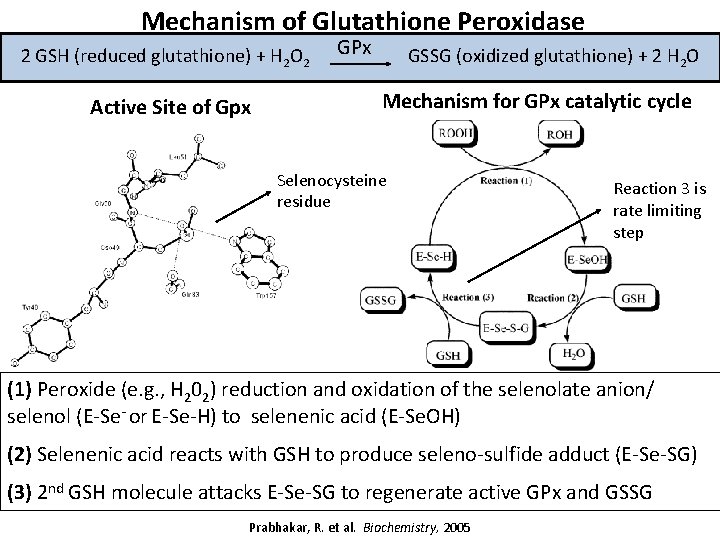
Mechanism of Glutathione Peroxidase 2 GSH (reduced glutathione) + H 2 O 2 Active Site of Gpx GPx GSSG (oxidized glutathione) + 2 H O 2 Mechanism for GPx catalytic cycle Selenocysteine residue Reaction 3 is rate limiting step (1) Peroxide (e. g. , H 202) reduction and oxidation of the selenolate anion/ selenol (E-Se- or E-Se-H) to selenenic acid (E-Se. OH) (2) Selenenic acid reacts with GSH to produce seleno-sulfide adduct (E-Se-SG) (3) 2 nd GSH molecule attacks E-Se-SG to regenerate active GPx and GSSG Prabhakar, R. et al. Biochemistry, 2005

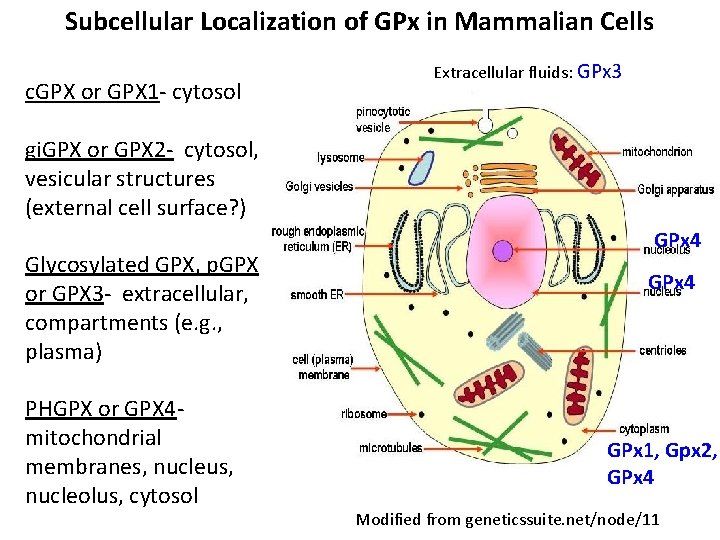
Subcellular Localization of GPx in Mammalian Cells c. GPX or GPX 1 - cytosol Extracellular fluids: GPx 3 gi. GPX or GPX 2 - cytosol, vesicular structures (external cell surface? ) Glycosylated GPX, p. GPX or GPX 3 - extracellular, compartments (e. g. , plasma) PHGPX or GPX 4 - mitochondrial membranes, nucleus, nucleolus, cytosol GPx 4 GPx 1, Gpx 2, GPx 4 Modified from geneticssuite. net/node/11

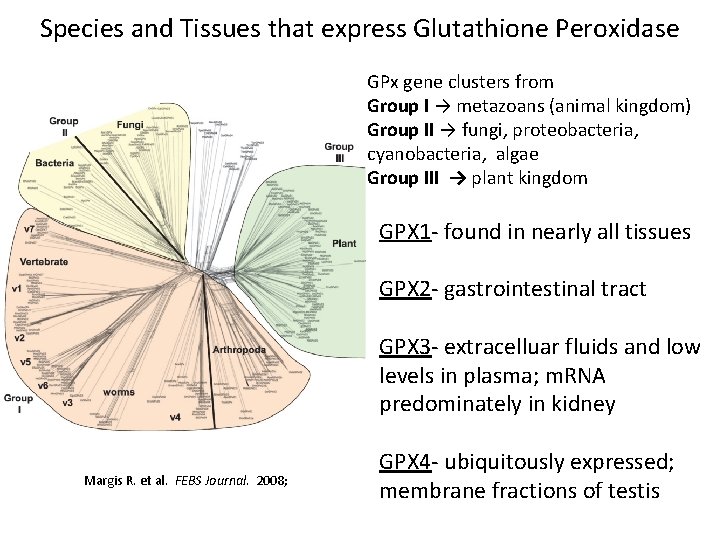
Species and Tissues that express Glutathione Peroxidase GPx gene clusters from Group I → metazoans (animal kingdom) Group II → fungi, proteobacteria, cyanobacteria, algae Group III → plant kingdom GPX 1 - found in nearly all tissues GPX 2 - gastrointestinal tract GPX 3 - extracelluar fluids and low levels in plasma; m. RNA predominately in kidney Margis R. et al. FEBS Journal. 2008; GPX 4 - ubiquitously expressed; membrane fractions of testis

Antioxidant Enzymes Beal, Nature. 2006 Oct 19; 443(7113): 787 -95.
 Lynda lytle holmstrom
Lynda lytle holmstrom Netoperacije 2
Netoperacije 2 Povijest austrije
Povijest austrije Qiang jiang
Qiang jiang Qiang jiang
Qiang jiang Cai guo qiang footprints of history
Cai guo qiang footprints of history Qiang yang hkust
Qiang yang hkust Qiang yang
Qiang yang Antioxidant classification
Antioxidant classification Vitamin e as antioxidant
Vitamin e as antioxidant Antioxidant system
Antioxidant system Antioxidant means
Antioxidant means Antioxidant chemical structure
Antioxidant chemical structure Antioxidant graphite electrode
Antioxidant graphite electrode Antioxidant free radical
Antioxidant free radical Oración a chiquitunga para pedir un milagro
Oración a chiquitunga para pedir un milagro Xiaolan zhang fordham
Xiaolan zhang fordham Zhang wang leukemia
Zhang wang leukemia Shengyu zhang
Shengyu zhang Dr ming zhang
Dr ming zhang Zhang et al 2014
Zhang et al 2014 Bitcoin
Bitcoin Daya zhang
Daya zhang Hongwei zhang
Hongwei zhang