ANTIFUNGAL DRUGS Modes of Action Mechanisms of Resistance































- Slides: 38

ANTIFUNGAL DRUGS Modes of Action Mechanisms of Resistance Sevtap Arikan, MD Hacettepe University Medical School Ankara Turkey

MOST COMMON FUNGAL PATHOGENS • Dermatophytes • Candida • Aspergillus • Cryptococcus • Rhizopus • . . .

ANTIFUNGAL DRUGS --by structure • POLYENES • • Amphotericin B, nystatin AZOLES Imidazoles: Ketoconazole. . Triazoles: Fluconazole, itraconazole, voriconazole, posaconazole, ravuconazole ALLYLAMINES Terbinafine, butenafine MORPHOLINE Amorolfine FLUORINATED PYRIMIDINE Flucytosine • ECHINOCANDINS Caspofungin, anidulafungin, micafungin • PEPTIDE-NUCLEOSIDE Nikkomycin Z • TETRAHYDROFURAN DERIVATIVES Sordarins, azasordarins • OTHER Griseofulvin

MODES of ACTION

ANTIFUNGAL DRUGS --by mode of action • Membrane disrupting • • • agents Amphotericin B, nystatin Ergosterol synthesis inhibitors Azoles, allylamines, morpholine Nucleic acid inhibitor Flucytosine Anti-mitotic (spindle disruption) Griseofulvin • Glucan synthesis inhibitors Echinocandins • Chitin synthesis inhibitor Nikkomycin • Protein synthesis inhibitors Sordarins, azasordarins

TARGETS for antifungal activity • Ergosterol (Cell membrane) ü Drug-ergosterol interaction Inhibition of ergosterol synthesis • RNA/EF 3 (Nucleic acid/protein synthesis) Incorporation of 5 -FU in RNA Inhibition of EF 3 • Glucan/Chitin (Cell wall) Inhibition of glucan/chitin synthesis

AMPHOTERICIN B generates pores in the membrane Clin Microbiol Rev 1999; 12: 501

TARGETS for antifungal activity • Ergosterol (Cell membrane) Drug-ergosterol interaction ü Inhibition of ergosterol synthesis • RNA/EF 3 (Nucleic acid/protein synthesis) Incorporation of 5 -FU in RNA Inhibition of EF 3 • Glucan/Chitin (Cell wall) Inhibition of glucan/chitin synthesis

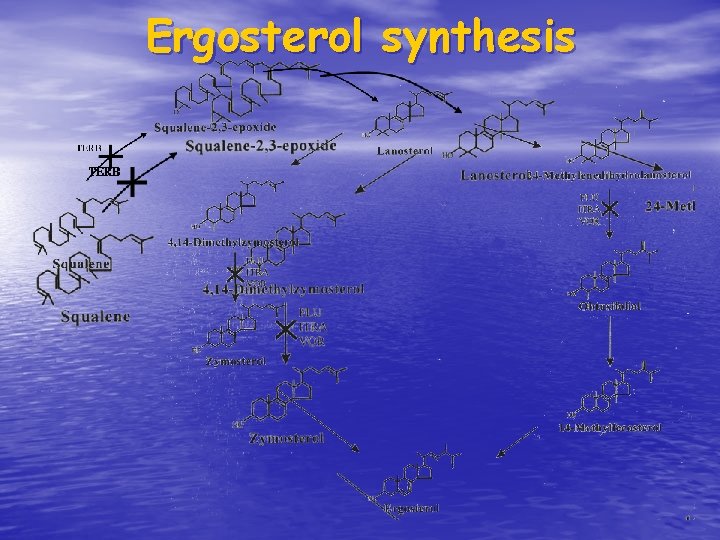
Ergosterol synthesis

Azoles, allylamines & morpholines inhibit specific ENZYMES Clin Microbiol Rev 1998; 11: 382

TARGETS for antifungal activity • Ergosterol (Cell membrane) Drug-ergosterol interaction Inhibition of ergosterol synthesis • RNA/EF 3 (Nucleic acid/Protein synthesis) ü Incorporation of 5 -FU into RNA Inhibition of EF 3 • Glucan/Chitin (Cell wall) Inhibition of glucan/chitin synthesis

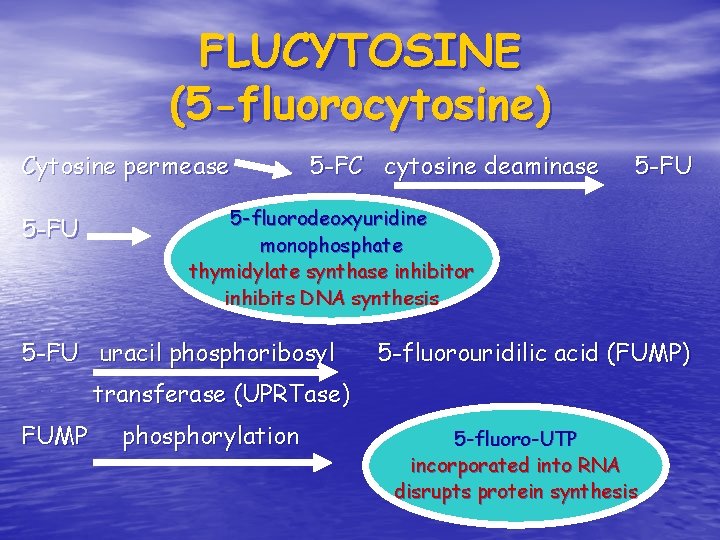
FLUCYTOSINE (5 -fluorocytosine) Cytosine permease 5 -FU 5 -FC cytosine deaminase 5 -FU 5 -fluorodeoxyuridine monophosphate thymidylate synthase inhibitor inhibits DNA synthesis 5 -FU uracil phosphoribosyl 5 -fluorouridilic acid (FUMP) transferase (UPRTase) FUMP phosphorylation 5 -fluoro-UTP incorporated into RNA disrupts protein synthesis

TARGETS for antifungal activity • Ergosterol (Cell membrane) Drug-ergosterol interaction Inhibition of ergosterol synthesis • RNA/EF 3 (Nucleic acid/protein synthesis) Incorporation of 5 -FU into RNA ü Inhibition of EF 3 • Glucan/Chitin (Cell wall) Inhibition of glucan/chitin synthesis

SORDARINS, AZASORDARINS • EF 3: A target in protein synthesis machinery unique to FUNGI • GM 237354. . . (sordarins) GW 471558. . . (azasordarins) • Yet investigational

TARGETS for antifungal activity • Ergosterol (Cell membrane) Drug-ergosterol interaction Inhibition of ergosterol synthesis • RNA/EF 3 (Nucleic acid/protein synthesis) Incorporation of 5 -FU into RNA Inhibition of EF 3 • Glucan/Chitin (Cell wall) üInhibition of glucan / chitin synthesis

ECHINOCANDINS Caspofungin is licensed • Inhibition of β-(1 -3) • • • glucan synthesis (of glucan synthase ? ? ) Secondary reduction in ergosterol & lanosterol Increase in chitin Kills hyphae at their growth tips and branching points Buds fail to seperate from the mother cell Yields osmotically sensitive fungal cells

TARGETS for antifungal activity • Ergosterol (Cell membrane) Drug-ergosterol interaction Inhibition of ergosterol synthesis • RNA/EF 3 (Nucleic acid/protein synthesis) Incorporation of 5 -FU into RNA Inhibition of EF 3 • Glucan/Chitin (Cell wall) ü Inhibition of glucan / chitin synthesis

NIKKOMYCIN • Competitive inhibition of chitin synthase • Yet investigational

MECHANISMS OF RESISTANCE

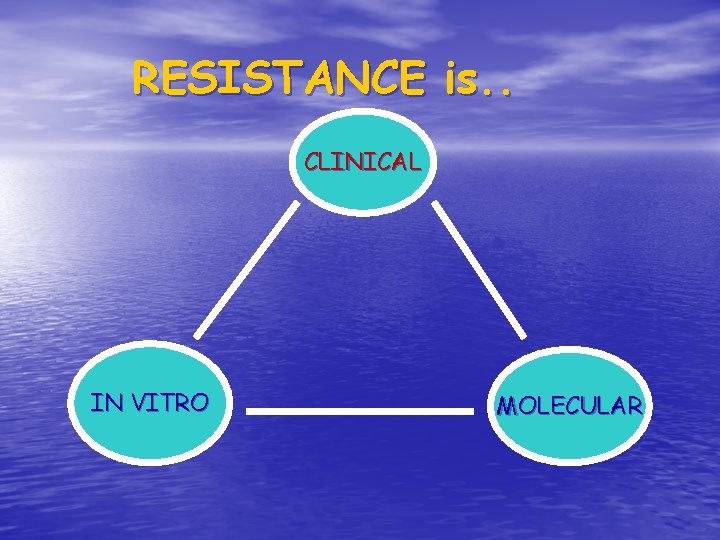
RESISTANCE is. . CLINICAL IN VITRO MOLECULAR

A resistant strain may be present due to: • • • Intrinsic resistance Replacement with a more resistant species Replacement with a more resistant strain Transient gene expressions that cause temporary resistance (epigenetic resistance) Alterations in cell type (? ) Genomic instability within a single strain (population bottleneck)

Clinical Resistance is a Multifactorial Issue • FUNGUS • HOST Immune status Site of infection Severity of infection Foreign devices Noncompliance with drug regimen Initial MIC Cell type: Yeast/hyphae. . Genomic stability Biofilm production Population bottlenecks • DRUG Fungistatic nature Dosing Pharmacokinetics Drug-drug interactions

Resistance to Amphotericin B • Technical difficulties in detection of • resistance in vitro In vivo resistance is rare C. lusitaniae, C. krusei C. neoformans Trichosporon spp. A. terreus S. apiospermum Fusarium spp. .

Mechanisms of Amphotericin B Resistance • Reduced ergosterol content (defective ERG 2 • • • or ERG 3 genes) Alterations in sterol content (fecosterol, episterol: reduced affinity) Alterations in sterol to phospholipid ratio Reorientation or masking of ergosterol Stationary growth phase Previous exposure to azoles (? )

Resistance to Azoles • • • Well-known particularly for fluconazole Data available also for other azoles A significant clinical problem RESISTANCE TO FLUCONAZOLE PRIMARY C. krusei Aspergillus C. glabrata C. norvegensis. . . SECONDARY C. albicans C. dubliniensis. . .

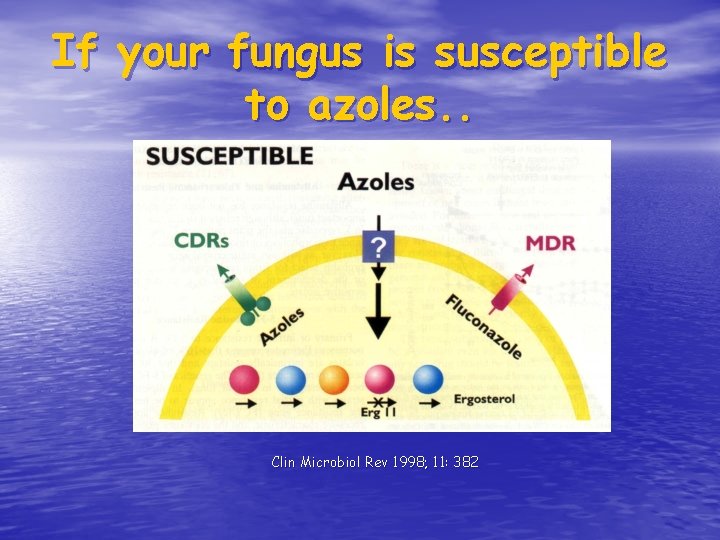
Mechanisms of Resistance to Azoles • Alteration of lanosterol (14 -alpha) demethylase • Overexpression of lanosterol demethylase • Energy-dependent efflux systems a. Major facilitator superfamily (MFS) proteins (BENr =MDR 1 of Candida. . . ) b. ATP-binding cassette (ABC) superfamily proteins (MDR, CDR of Candida) • Changes in sterol and/or phospholipid composition of fungal cell membrane (decreased permeability)

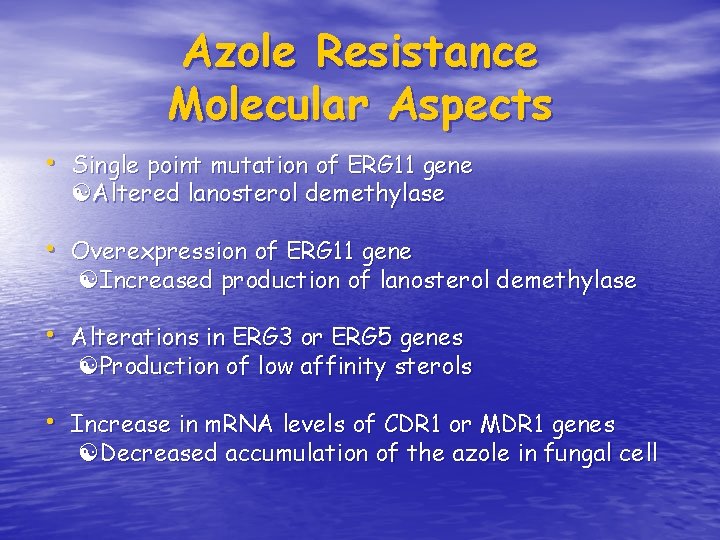
Azole Resistance Molecular Aspects • Single point mutation of ERG 11 gene Altered lanosterol demethylase • Overexpression of ERG 11 gene Increased production of lanosterol demethylase • Alterations in ERG 3 or ERG 5 genes Production of low affinity sterols • Increase in m. RNA levels of CDR 1 or MDR 1 genes Decreased accumulation of the azole in fungal cell

If your fungus is susceptible to azoles. . Clin Microbiol Rev 1998; 11: 382

If it is azole-resistant. . Clin Microbiol Rev 1998; 11: 382

Secondary Resistance in C. albicans to Fluconazole CID 1997; 25: 908 -910

Resistance to Terbinafine • Very rare • Primary resistance to terbinafine in a T. rubrum strain (ICAAC 2001, abst. no. J-104) • Mechanism: (? ) CDR 1 -mediated efflux (possible)

Resistance to Flucytosine • PRIMARY non-albicans Candida C. neoformans Aspergillus (highest) • SECONDARY C. albicans C. neoformans Secondary resistance develops following flucytosine MONOtherapy.

Mechanisms of Resistance to Flucytosine • Loss of permease activity • Loss of cytosine deaminase activity • Decrease in the activity of UPRTase

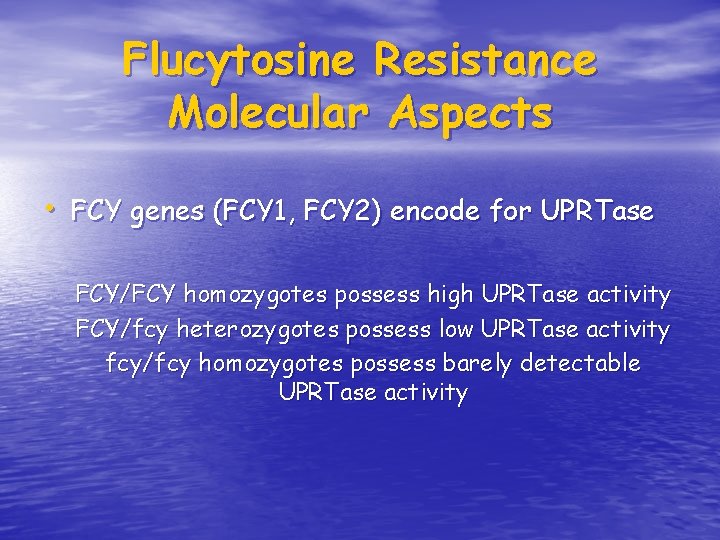
Flucytosine Resistance Molecular Aspects • FCY genes (FCY 1, FCY 2) encode for UPRTase FCY/FCY homozygotes possess high UPRTase activity FCY/fcy heterozygotes possess low UPRTase activity fcy/fcy homozygotes possess barely detectable UPRTase activity

Resistance to Echinocandins PRIMARY C. neoformans Fusarium spp. SECONDARY (? ) The only licensed member is caspofungin (Jan 2001, USA). Resistant mutants due to therapy are not available.

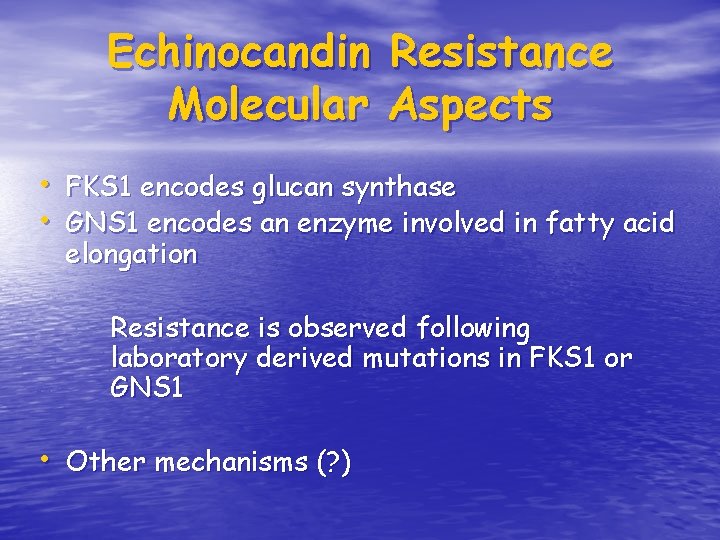
Echinocandin Molecular Resistance Aspects • FKS 1 encodes glucan synthase • GNS 1 encodes an enzyme involved in fatty acid elongation Resistance is observed following laboratory derived mutations in FKS 1 or GNS 1 • Other mechanisms (? )

Future Directions to Avoid Development of Resistance • Proper dosing strategies • Restricted and well-defined indications for prophylaxis with azoles Fungi will continue to develop NEW resistance mechanisms!. .

Final word • Antifungal resistance is a complex, gradual and multifactorial issue • Several uncertainties remain • Molecular assays to detect resistance are not simple • The best way to improve the efficacy of antifungal therapy is to improve the immune status of the host
 Antifungal tablets
Antifungal tablets Classification of antifungal drugs
Classification of antifungal drugs Mechanism of antifungal drugs
Mechanism of antifungal drugs Antifungal drugs classification
Antifungal drugs classification Resistance mechanisms of bacteria
Resistance mechanisms of bacteria Antifungal sensitivity test
Antifungal sensitivity test Canesten solution boots
Canesten solution boots Friction and air resistance
Friction and air resistance The inlet pressure in a constant rate filtration
The inlet pressure in a constant rate filtration Aspirin mechanism of action
Aspirin mechanism of action Mechanism of action of asprin
Mechanism of action of asprin Antiepileptic drugs classification
Antiepileptic drugs classification Fibrinolysis mechanism
Fibrinolysis mechanism Anticholinergic drugs mechanism of action
Anticholinergic drugs mechanism of action Classes of antiarrhythmic drugs
Classes of antiarrhythmic drugs Thrombolytic drugs mechanism of action
Thrombolytic drugs mechanism of action Atropine receptor
Atropine receptor Antiarrhythmic drugs mechanism of action
Antiarrhythmic drugs mechanism of action Thrombolytic drugs mechanism of action
Thrombolytic drugs mechanism of action Emetrol mechanism of action
Emetrol mechanism of action Parasympathomimetic drugs
Parasympathomimetic drugs Dromotropic effect
Dromotropic effect Mechanism of action of antiepileptic drugs
Mechanism of action of antiepileptic drugs Insecticide resistance action committee
Insecticide resistance action committee Exposition in a rose for emily
Exposition in a rose for emily Medias res
Medias res Suit the action to the word the word to the action meaning
Suit the action to the word the word to the action meaning Exposition rising action climax
Exposition rising action climax Stages of plot development
Stages of plot development What are discourse communities
What are discourse communities Service delivery mechanism
Service delivery mechanism Mechanisms of heat loss in newborn
Mechanisms of heat loss in newborn Youjip
Youjip What are the mechanisms of evolution
What are the mechanisms of evolution Defense mechanisms nursing
Defense mechanisms nursing Ego defense mechanisms in lord of the flies
Ego defense mechanisms in lord of the flies Ego defenses
Ego defenses E-commerce mechanisms definition
E-commerce mechanisms definition Defense mechanisms nursing
Defense mechanisms nursing